Donor-recipient mismatch at the SIRPA locus adversely affects kidney allograft outcomes
- PMID: 40668892
- PMCID: PMC12331381
- DOI: 10.1126/scitranslmed.ady1135
Donor-recipient mismatch at the SIRPA locus adversely affects kidney allograft outcomes
Abstract
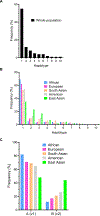
Donor-recipient mismatches in histocompatibility antigens recognized by lymphoid cells have been demonstrated to adversely affect allograft outcomes. In contrast, it remains unclear whether mismatches sensed by innate myeloid cells have a similar effect. We investigated the consequences of mismatch in the polymorphic gene encoding signal regulatory protein α (SIRPα) on kidney allograft pathology and survival in mice and humans. We found that SIRPα variants elicit monocyte activation by binding to CD47 and that eliminating SIRPα mismatch or recipient CD47 expression prevented chronic allograft pathology in mice receiving major histocompatibility complex (MHC)-mismatched renal allografts. Human genomic analysis identified two haplotype categories, A and B, encoding SIRPα variants with distinct CD47 binding interfaces. In kidney transplant recipients (N = 455), SIRPα mismatch was associated with increased acute rejection and graft fibrosis in the first posttransplant year, and A recipients of B kidneys had reduced long-term graft survival (hazard ratio, 3.2; 95% confidence interval, 1.5 to 6.9; P = 0.002), a finding that was confirmed in an independent validation cohort (N = 258). Moreover, monocytes in these graft recipients had an activated phenotype. The effects of SIRPα mismatch were independent of ancestry, human leukocyte antigen mismatch, donor-specific antibodies, and delayed graft function. Therefore, these data demonstrate that a donor-recipient mismatch that causes innate immune activation is a determinant of kidney transplantation outcomes.
Conflict of interest statement
Figures






References
-
- Callemeyn J, Lamarthee B, Koenig A, Koshy P, Thaunat O, Naesens M, Allorecognition and the spectrum of kidney transplant rejection. Kidney Int. 101, 692–710 (2022). - PubMed
-
- Chow KV, Delconte RB, Huntington ND, Tarlinton DM, Sutherland RM, Zhan Y, Lew AM, Innate allorecognition results in rapid accumulation of monocyte-derived dendritic cells. J. Immunol. 197, 2000–2008 (2016). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

