Modulating immune cell fate and inflammation through CRISPR-mediated DNA methylation editing
- PMID: 40668918
- PMCID: PMC12266127
- DOI: 10.1126/sciadv.adt1644
Modulating immune cell fate and inflammation through CRISPR-mediated DNA methylation editing
Abstract
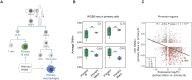
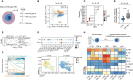
Immune cell differentiation and activation are associated with widespread DNA methylation changes; however, the causal relationship between these changes and their impact in shaping cell fate decisions still needs to be fully elucidated. Here, we conducted a genome-wide analysis to investigate the relationship between DNA methylation and gene expression at gene regulatory regions in human immune cells. By using CRISPR-dCas9-TET1 and -DNMT3A epigenome editing tools, we successfully established a cause-and-effect relationship between the DNA methylation levels of the promoter of the interleukin-1 receptor antagonist (IL1RN) gene and its expression. We observed that modifying the DNA methylation status of the IL1RN promoter is sufficient to alter human myeloid cell fate and change the cellular response to inflammatory and pathogenic stimuli. Collectively, our findings demonstrate the potential of targeting specific DNA methylation events to directly modulate immune and inflammatory responses, providing a proof of principle for intervening in a broad range of inflammation-related diseases.
Figures



References
-
- Bird A., DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 (2002). - PubMed
-
- Charlton J., Jung E. J., Mattei A. L., Bailly N., Liao J., Martin E. J., Giesselmann P., Brändl B., Stamenova E. K., Müller F.-J., Kiskinis E., Gnirke A., Smith Z. D., Meissner A., TETs compete with DNMT3 activity in pluripotent cells at thousands of methylated somatic enhancers. Nat. Genet. 52, 819–827 (2020). - PMC - PubMed
-
- Dai H.-Q., Wang B.-A., Yang L., Chen J.-J., Zhu G.-C., Sun M.-L., Ge H., Wang R., Chapman D. L., Tang F., Sun X., Xu G.-L., TET-mediated DNA demethylation controls gastrulation by regulating Lefty–Nodal signalling. Nature 538, 528–532 (2016). - PubMed
-
- Greve G., Andrieux G., Schlosser P., Blagitko-Dorfs N., Rehman U.-U., Ma T., Pfeifer D., Heil G., Neubauer A., Krauter J., Heuser M., Salih H. R., Döhner K., Döhner H., Hackanson B., Boerries M., Lübbert M., In vivo kinetics of early, non-random methylome and transcriptome changes induced by DNA-hypomethylating treatment in primary AML blasts. Leukemia 37, 1018–1027 (2023). - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

