Guardian of myelin and neural Integrity: foxo1a through slc7a11 mitigating oxidative damage in myelin
- PMID: 40669207
- PMCID: PMC12283898
- DOI: 10.1016/j.redox.2025.103763
Guardian of myelin and neural Integrity: foxo1a through slc7a11 mitigating oxidative damage in myelin
Abstract
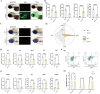
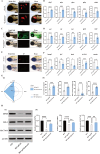
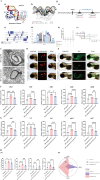
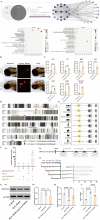
The emergence of myelin marks an evolutionary leap from jawless to jawed vertebrates. Although myelin's role in promoting rapid neural signal transmission and brain complexity is known, its neuroprotective mechanisms in complex signal transmission remain unclear. This study identifies the critical FoxO gene family member, foxo1a, as essential to the evolution of jawed vertebrates by comparing divergence times and gene family heterogeneity between jawless and jawed vertebrates. We found that foxo1a is located in zebrafish oligodendrocytes and myelin, playing a key antioxidant protective role. Specifically, we found that knocking out the foxo1a gene leads to abnormal myelin development in the central nervous system of zebrafish, a reduction in oligodendrocytes, astrocytes, and myelin markers, and induces freezing behavior. Further research revealed that this is related to oxidative stress responses and ferroptosis in the central nervous system of zebrafish following the deficiency of the foxo1a gene. Mechanistically, we discovered that foxo1a is involved in regulating oxidative stress responses and iron homeostasis in the central nervous system by directly regulating the promoter activity of the slc7a11 gene. In terms of application, we found that exogenous supplementation of foxo1a can exert antioxidant protective effects in a copper sulfate-induced myelin damage model. More importantly, we found a parallelism of the foxo1a-slc7a11 axis in both zebrafish and human cells, suggesting that the foxo1a-slc7a11 axis might be an evolutionarily conserved neural defense strategy in jawed vertebrates. In conclusion, our study elucidates the critical role of foxo1a in maintaining antioxidant homeostasis in the central nervous system and provides new insights into the adaptive evolution of the central nervous system in jawed vertebrates.
Keywords: Antioxidant defense; Ferroptosis; Jawed vertebrates; Myelin; foxo1a.
Copyright © 2025. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors have declared that no conflict of interest exists.
Figures



Similar articles
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
MiR-370-3p regulate TLR4/SLC7A11/GPX4 to alleviate the progression of glucocorticoids-induced osteonecrosis of the femoral head by promoting osteogenesis and suppressing ferroptosis.J Orthop Translat. 2025 Feb 12;51:337-358. doi: 10.1016/j.jot.2024.10.014. eCollection 2025 Mar. J Orthop Translat. 2025. PMID: 40584015 Free PMC article.
-
Intestinal inflammation and microbiota modulation impact cochlear function: emerging insights in gut-ear axis.Cell Commun Signal. 2025 Jul 26;23(1):357. doi: 10.1186/s12964-025-02338-1. Cell Commun Signal. 2025. PMID: 40713718 Free PMC article.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
-
Comparison of self-administered survey questionnaire responses collected using mobile apps versus other methods.Cochrane Database Syst Rev. 2015 Jul 27;2015(7):MR000042. doi: 10.1002/14651858.MR000042.pub2. Cochrane Database Syst Rev. 2015. PMID: 26212714 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

