Elaboration of molecular glues that target TRIM21 into TRIMTACs that degrade protein aggregates
- PMID: 40670348
- PMCID: PMC12267526
- DOI: 10.1038/s41467-025-61818-7
Elaboration of molecular glues that target TRIM21 into TRIMTACs that degrade protein aggregates
Abstract
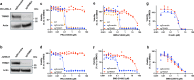
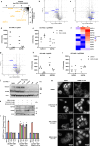
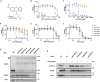
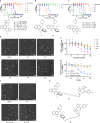
Approaches for the discovery of molecular glues remain limited. Here we report a phenotypic screening approach in which cytotoxins whose mechanisms require ubiquitination show a gain of viability following pharmacological inhibition of the Ubiquitin-like modifier activating enzyme (UBA1/UAE). This approach reveals PRLX-93936 and BMS-214662 as molecular glues that directly target the E3 ligase TRIM21 to induce degradation of nucleoporin proteins and inhibit nuclear trafficking. The cytotoxicity of these agents correlates strongly with TRIM21 expression, suggesting re-evaluation of these clinically tested agents in patients with TRIM21-high cancers. Relative to recently disclosed TRIM21-targeting glues, PRLX-93936 and newly-synthesized analogs represent a distinct structural series, lack known cellular off-targets, and offer greatly enhanced potency. Additionally, we elaborate PRLX-93936 to a heterobifunctional degrader that uses wild-type TRIM21 to degrade a multimeric protein. Together, our work creates opportunities for targeted protein degradation and enables the design of additional TRIM21-targeting glues and Proteolysis-Targeting Chimeras (PROTACs).
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.J.A. and M.A.S.dG. are inventors on a provisional patent application (63/735,365) filed by Case Western Reserve University on the synthesis of PRLX-93936 analogs and their applications towards degrading protein aggregates. The other authors declare no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

