A conserved motif within the NSP2 of SARS-CoV-2 is required for processing of the distal NSP1/NSP2 junction by NSP3
- PMID: 40670498
- PMCID: PMC12267441
- DOI: 10.1038/s41598-025-10244-2
A conserved motif within the NSP2 of SARS-CoV-2 is required for processing of the distal NSP1/NSP2 junction by NSP3
Abstract
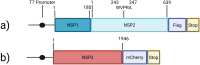
In 2019, the severe acute respiratory syndrome coronavirus 2 virus (SARS-CoV-2) started to spread globally and caused the COVID-19 pandemic. SARS-CoV-2, like other members of the Coronaviridae, has a single-stranded, positive sense RNA genome about 30 kb in length, which is translated to generate 16 non-structural proteins (NSPs); a set of sub-genomic mRNAs encode the structural and accessory proteins. The ORF1a precursor includes NSP1-11 and is processed by virus-encoded proteases to produce the mature proteins. We recently identified a short, highly conserved motif (YCPRP) within the structural protein precursor of foot-and-mouth disease virus (FMDV), a member of the Picornaviridae. This motif is conserved among picornaviruses and is found as (W/F/Y)-x-P-R-(P/A). The motif has a major influence on the processing of the FMDV capsid precursor (P1-2A) by the viral protease 3Cpro. We have now identified a similar motif (WVPRA) within the NSP2 of SARS-CoV-2. Interestingly, this motif is required for the efficient processing of the NSP1-NSP2 junction by the SARS-CoV-2 protease PLpro (NSP3) and a single amino acid substitution within the motif can abrogate cleavage of this junction. We hypothesise that this motif acts, within NSP1-NSP2, to enable this precursor to fold correctly and allow efficient processing of the NSP1/NSP2 junction.
Keywords: Coronavirus; Intramolecular chaperone; NSP2; Proteolytic processing; SARS-CoV-2.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Viral determinants of cis- and trans-cleavage by SARS-CoV-2 Nsp3 and an on-off reporter for monitoring intracellular protease activity.Antiviral Res. 2025 Oct;242:106262. doi: 10.1016/j.antiviral.2025.106262. Epub 2025 Aug 18. Antiviral Res. 2025. PMID: 40835029
-
Structural and functional analyses of SARS-CoV-2 Nsp3 and its specific interactions with the 5' UTR of the viral genome.Microbiol Spectr. 2025 Aug 5;13(8):e0287124. doi: 10.1128/spectrum.02871-24. Epub 2025 Jul 7. Microbiol Spectr. 2025. PMID: 40621910 Free PMC article.
-
Structure-function mapping and mechanistic insights on the SARS CoV2 Nsp1.Protein Sci. 2024 Dec;33(12):e5228. doi: 10.1002/pro.5228. Protein Sci. 2024. PMID: 39584680
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

