Expression of DNAJB1-PRKACA oncogene suppresses the differentiation potential of liver progenitor organoids towards a hepatocyte lineage
- PMID: 40670531
- PMCID: PMC12267505
- DOI: 10.1038/s41598-025-11028-4
Expression of DNAJB1-PRKACA oncogene suppresses the differentiation potential of liver progenitor organoids towards a hepatocyte lineage
Abstract
Fibrolamellar carcinoma (FLC) is a type of primary liver cancer that predominantly affects healthy adolescents and young adults in a background of normal liver. The DNAJB1-PRKACA fusion gene is an oncogenic driver in FLC tumors. To investigate the oncogenic mechanisms of this fusion gene, we developed a model using human liver progenitor organoids engineered to express DNAJB1-PRKACA. Single-nucleus RNA sequencing of these organoids revealed an upregulation of genes that significantly overlap with those expressed in FLC epithelial cells. Additionally, the expression of DNAJB1-PRKACA led to the downregulation of genes coding for markers of mature epithelial cells, indicating a shift toward a less differentiated state. When compared to wild-type liver progenitor organoids, which exhibit a strong ability to differentiate into hepatocytes, the DNAJB1-PRKACA-expressing liver progenitor organoids displayed a markedly reduced capacity for hepatocyte differentiation. These findings suggest that the DNAJB1-PRKACA fusion gene disrupts the normal differentiation process of liver progenitor cells.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures
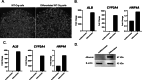



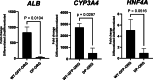
Similar articles
-
A novel high-throughput screening platform to identify inhibitors of DNAJB1-PRKACA-driven transcriptional activity in fibrolamellar carcinoma.SLAS Discov. 2025 Apr;32:100221. doi: 10.1016/j.slasd.2025.100221. Epub 2025 Feb 11. SLAS Discov. 2025. PMID: 39947627 Free PMC article.
-
CRISPR/Cas9 Engineering of Adult Mouse Liver Demonstrates That the Dnajb1-Prkaca Gene Fusion Is Sufficient to Induce Tumors Resembling Fibrolamellar Hepatocellular Carcinoma.Gastroenterology. 2017 Dec;153(6):1662-1673.e10. doi: 10.1053/j.gastro.2017.09.008. Epub 2017 Sep 18. Gastroenterology. 2017. PMID: 28923495 Free PMC article.
-
Increased Protein Kinase A Activity Induces Fibrolamellar Hepatocellular Carcinoma Features Independent of DNAJB1.Cancer Res. 2024 Aug 15;84(16):2626-2644. doi: 10.1158/0008-5472.CAN-23-4110. Cancer Res. 2024. PMID: 38888469 Free PMC article.
-
How Do I Diagnose Fibrolamellar Carcinoma?Mod Pathol. 2025 Apr;38(4):100711. doi: 10.1016/j.modpat.2025.100711. Epub 2025 Jan 13. Mod Pathol. 2025. PMID: 39814265 Review.
-
Fibrolamellar Hepatocellular Carcinoma: Mechanistic Distinction From Adult Hepatocellular Carcinoma.Pediatr Blood Cancer. 2016 Jul;63(7):1163-7. doi: 10.1002/pbc.25970. Epub 2016 Mar 14. Pediatr Blood Cancer. 2016. PMID: 26990031 Free PMC article. Review.
Cited by
-
Does Chemotherapy Have an Effect on the Treatment Success of Children and Adolescents with Unresectable Hepatocellular Carcinoma? Findings from the German Liver Tumour Registry.Cancers (Basel). 2025 Jul 23;17(15):2444. doi: 10.3390/cancers17152444. Cancers (Basel). 2025. PMID: 40805147 Free PMC article.
References
-
- O’Neill, A. F. et al. Fibrolamellar carcinoma: an entity all its own. Curr Probl. Cancer Aug. 45 (4), 100770. 10.1016/j.currproblcancer.2021.100770 (2021). - PubMed
-
- Weeda, V. B. et al. Fibrolamellar variant of hepatocellular carcinoma does not have a better survival than conventional hepatocellular carcinoma–results and treatment recommendations from the childhood liver tumour strategy group (SIOPEL) experience. Eur J. Cancer Aug. 49 (12), 2698–2704. 10.1016/j.ejca.2013.04.012 (2013). - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

