Mitochondrial respiration is necessary for CD8+ T cell proliferation and cell fate
- PMID: 40670617
- PMCID: PMC12307223
- DOI: 10.1038/s41590-025-02202-x
Mitochondrial respiration is necessary for CD8+ T cell proliferation and cell fate
Abstract
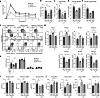
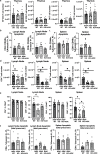
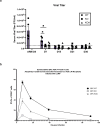
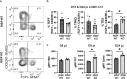
Mitochondrial electron transport chain (ETC) function is linked to the generation of ATP, signaling molecules including reactive oxygen species (ROS), pyrimidines and tricarboxylic acid cycle metabolites1. Mitochondrial electron transport is required for T cell proliferation2-4. However, which mitochondrial ETC functions are necessary for each dynamic state of CD8+ T cell responses is unknown. Here we report that impairing mitochondrial complex III function, which diminishes respiration, proton pumping linked to ATP production and superoxide production, decreases peripheral naive numbers, antigen-induced CD8+ T cell proliferation and memory formation. Acute stimulation of mitochondrial complex III-deficient CD8+ T cells induced an exhausted-like phenotype. Expression of Ciona intestinalis alternative oxidase (AOX) in mitochondrial complex III-deficient CD8+ T cells restores respiration without generating ROS or proton pumping, and rescues proliferation and the exhausted phenotype but not naive or memory formation. Thus, T cell development, proliferation and memory formation have distinct requirements for mitochondrial complex III ROS.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: M.S. is a shareholder in a start-up company founded to develop therapeutics based on AOX. The other authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
- 75N92020D00005/HL/NHLBI NIH HHS/United States
- F30CA250236/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 75N93023D00005/AI/NIAID NIH HHS/United States
- Cancer Research Institute Fellow/Cancer Research Institute (CRI)
- P01 HL154998/HL/NHLBI NIH HHS/United States
- 75N95020D00005/DA/NIDA NIH HHS/United States
- 5P01HL154998/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 75N99020D00005/OF/ORFDO NIH HHS/United States
- R01AI148190/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 75N93022D00005/AI/NIAID NIH HHS/United States
- F30 CA250236/CA/NCI NIH HHS/United States
- R01 AI148190/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

