The proteostatic landscape of healthy human oocytes
- PMID: 40670770
- PMCID: PMC12361380
- DOI: 10.1038/s44318-025-00493-2
The proteostatic landscape of healthy human oocytes
Abstract
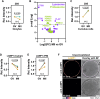
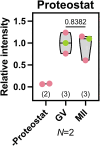
Oocytes, female germ cells that develop into eggs, are among the longest-lived cells in the animal body. Recent studies on mouse oocytes highlight unique adaptations in protein homeostasis (proteostasis) within these cells. However, the mechanisms of proteostasis in human oocytes remain virtually unstudied. We present the first large-scale study of proteostatic activity in human oocytes using over 100 freshly donated oocytes from 21 healthy women aged 19-34 years. We analysed the activity and distribution of lysosomes, proteasomes, and mitochondria in both immature and mature oocytes. Notably, human oocytes exhibit nearly twofold lower proteolytic activity than surrounding somatic cells, with further decreases as oocytes mature. Oocyte maturation is also coupled with lysosomal exocytosis and a decrease in mitochondrial membrane potential. We propose that reduced organelle activity preserves key cellular components critical for early embryonic development during the prolonged maturation of human oocytes. Our findings highlight the distinctive biology of human oocytes and the need to investigate human-specific reproductive biology to address challenges in female fertility.
Keywords: Female Fertility; Human Oocytes; Lysosomes; Mitochondria; Proteostasis.
© 2025. The Author(s).
Conflict of interest statement
Disclosure and competing interests statement. The authors declare no competing interests.
Figures







References
-
- Berkers CR, Leeuwen FWB, van, Groothuis TA, Peperzak V, Tilburg EW van, Borst J, Neefjes JJ, Ovaa H (2007) Profiling proteasome activity in tissue with fluorescent probes. Mol Pharm 4:739–748 - PubMed
-
- Chazotte B (2011) Labeling lysosomes in live cells with LysoTracker. Cold Spring Harb Protoc 2011:pdb.prot5571 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

