Campylobacter jejuni regulates cell cycle progression to potentiate host cell invasion
- PMID: 40671100
- PMCID: PMC12269301
- DOI: 10.1186/s12964-025-02348-z
Campylobacter jejuni regulates cell cycle progression to potentiate host cell invasion
Abstract
Background: Campylobacter jejuni is associated with enteritis in humans and domestic animals. Acute C. jejuni-mediated enteritis requires bacterial invasion of intestinal cells with an ensuing host inflammatory response. Known is that C. jejuni invasion of human epithelial cells is dependent on host cell-focal adhesion components, which link the extracellular matrix to the actin cytoskeleton of a cell. Based on the observation that C. jejuni cell invasion engages many of the same components involved in regulating the cell cycle, we hypothesized that C. jejuni regulates the host cell cycle.
Methods: Flow cytometry was used to detect the cell cycle phases (G1, S, G2 and M). Single-cell RNA-sequencing (scRNA-seq) and reverse transcriptase quantitative PCR (RT-qPCR) were used to determine the differential gene expressions of uninfected and C. jejuni-infected cells. Infection assays and confocal microscopy were employed to determine the rate of bacterial invasion and intracellular localization of C. jejuni-infected cells. Quantification of Interleukin-8 (IL-8) was determined by the ELISAs.
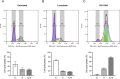
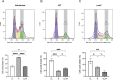
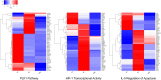
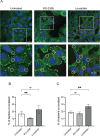
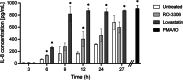
Results: INT 407 cells infected with C. jejuni showed a slower rate of cell cycle progression and a greater percentage of cells in the G1 cell cycle phase. scRNA-seq and RT-qPCR analysis of C. jejuni-infected cells corroborated the result, revealing host genes responsive to C. jejuni infection, including genes associated with cell cycle regulation, focal adhesions, inflammatory cytokines, and oxidative stress. Cell cycle synchronization coupled with the gentamicin-protection revealed that C. jejuni preferentially invades cells in the G1 phase. Moreover, an increase was observed in the number of bacteria colocalized with paxillin, a critical component of focal adhesion complexes, during the G1 phase. The infection of INT 407 cells in the G1 phase also increased the secretion of the proinflammatory cytokine IL-8 from cells.
Conclusions: Based on the data, we propose that acute C. jejuni-mediated enteritis (campylobacteriosis) alters the cell cycle phase of enterocytes, cytokine production, and immune cell recruitment, disrupting the intestinal permeability barrier.
Keywords: Cell cycle; Cell invasion; Inflammatory response; Oxidative stress; scRNA-seq.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures






Similar articles
-
Assessing Campylobacter jejuni Extracellular Vesicle-Host Interaction Using a Microfluidic Platform with Caco-2 Spheroides-on-Chip.ACS Biomater Sci Eng. 2025 Aug 11;11(8):4818-4829. doi: 10.1021/acsbiomaterials.5c00750. Epub 2025 Jun 23. ACS Biomater Sci Eng. 2025. PMID: 40548493
-
Exposure of broiler chickens to deoxynivalenol and Campylobacter jejuni induces substantial changes in intestinal gene expression.Sci Rep. 2025 Apr 19;15(1):13531. doi: 10.1038/s41598-025-97672-2. Sci Rep. 2025. PMID: 40253521 Free PMC article.
-
Transcriptome-wide N6-methyladenosinem modifications analysis of chicken cecum in responding to Campylobacter jejuni inoculation.Front Immunol. 2025 Jul 30;16:1630008. doi: 10.3389/fimmu.2025.1630008. eCollection 2025. Front Immunol. 2025. PMID: 40808945 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
References
-
- Berridge MJ. Cell signalling biology: module 9 - cell cycle and proliferation. Biochem J. 2012;6. 10.1042/csb0001009.
-
- Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. BioEssays. 1995;17(6):471–80. 10.1002/bies.950170603. (PubMed PMID: 7575488). - PubMed
-
- Nougayrede JP, Taieb F, De Rycke J, Oswald E. Cyclomodulins: bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 2005;13(3):103–10. 10.1016/j.tim.2005.01.002. (PubMed PMID: 15737728). - PubMed
-
- Oswald E, Nougayrede JP, Taieb F, Sugai M. Bacterial toxins that modulate host cell-cycle progression. Curr Opin Microbiol. 2005;8(1):83–91. 10.1016/j.mib.2004.12.011. (PubMed PMID: 15694861). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

