Genetic mutations disrupt the coordinated mode of tyrosinase's intra-melanosomal domain
- PMID: 40671279
- PMCID: PMC12267667
- DOI: 10.1002/pro.70209
Genetic mutations disrupt the coordinated mode of tyrosinase's intra-melanosomal domain
Abstract
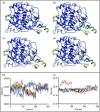
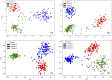
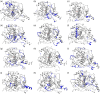
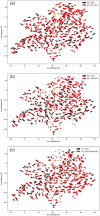
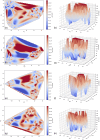
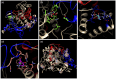
Oculocutaneous albinism type 1 is a genetic disorder caused by the disruption of tyrosinase activity in the melanogenesis pathway. The tyrosinase's intramelanosomal domain can be subdivided into the catalytic and Cys-rich subdomains, integral for protein stability and catalytic activity. To understand the movement in the tyrosinase intra-melanosomal subdomains and their link to its catalytic activity, we perform essential dynamics on homology models for tyrosinase and the mutant variants R217Q, R402Q, and R217Q/R402Q. Dimensional reduction techniques, such as principal component analysis (PCA), are fundamental to systematically comprehending collective movements in protein structure. The alpha-carbon atomic coordinates for all residues across a 100-ns molecular dynamics trajectory were input into the PCA function, and the results were analyzed alongside correlated movements and free energy profiles for each protein structure. The PCA-identified coordinated movement underlying the stable conformations of wild-type tyrosinase arises within the H9 and H10 helices, which are proximal to the flexible tunnel system and the interface of the catalytic and Cys-rich subdomains. In contrast, genetic mutations R217Q and R217Q/R402Q disrupt the coordinated movement of the tyrosinase intra-melanosomal domain, indicating a cause of mutant variant instability.
Keywords: PCA; coordinated protein motions; molecular dynamics; oculocutaneous albinism type 1; tyrosinase.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures



Update of
-
Genetic mutations disrupt the coordinated mode of tyrosinase intra-melanosomal domain.bioRxiv [Preprint]. 2025 Apr 25:2025.04.21.649833. doi: 10.1101/2025.04.21.649833. bioRxiv. 2025. Update in: Protein Sci. 2025 Aug;34(8):e70209. doi: 10.1002/pro.70209. PMID: 40568124 Free PMC article. Updated. Preprint.
References
-
- Abulaiti D, Tuerxun N, Wang H, Ma L, Zhao F, Liu Y, et al. Fungal secondary metabolites as a potential inhibitor of t315i‐ bcr::Abl1 mutant in chronic myeloid leukemia by molecular docking, molecular dynamics simulation and binding free energy exploration approaches. J Genet Eng Biotechnol. 2024;22(4):100444. - PMC - PubMed
-
- Amadei A, Linssen AB, Berendsen HJ. Essential dynamics of proteins. Proteins. 1993;17(4):412–425. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

