Splice-modulating antisense oligonucleotides targeting a pathogenic intronic variant in adult polyglucosan body disease correct mis-splicing and restore enzyme activity in patient cells
- PMID: 40671519
- PMCID: PMC12266137
- DOI: 10.1093/nar/gkaf658
Splice-modulating antisense oligonucleotides targeting a pathogenic intronic variant in adult polyglucosan body disease correct mis-splicing and restore enzyme activity in patient cells
Abstract
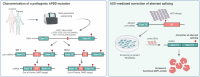
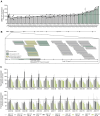
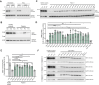
Adult polyglucosan body disease (APBD) is a rare, adult-onset neurodegenerative disorder caused by loss-of-function variants in the glycogen branching enzyme (GBE1) gene, essential for glycogen biosynthesis. The second most common pathogenic mutation in APBD (c.2053-3358_2053-3350delinsTGTTTTTTACATGACAGGT) is a deep intronic deletion insertion (indel) variant creating an ectopic splice acceptor site, resulting in a mutant transcript with a pseudoexon encoding an unstable truncated protein. Such mutations can be effectively targeted with splice-modulating antisense oligonucleotides (ASOs) to restore normal splicing. Here, we characterized the indel in-depth using long-read sequencing techniques and discovered several new features of the mutant transcript. The indel sequence varies from the previously identified sequence by a nucleotide, and the usage of the ectopic splice site results in two mutant isoforms, both of which are targets of cellular nonsense-mediated decay. High-throughput screening in patient-derived fibroblasts identified multiple lead candidates that effectively blocked the ectopic splice site and increased the canonical GBE1 transcript and protein. Functional analysis confirmed that treatment with the lead ASOs significantly improved GBE1 enzyme activity in patient cells, validating their therapeutic potential. Taken together, our data demonstrate the successful discovery of ASOs that correct mis-splicing, thus offering a promising treatment for a subset of APBD patients.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
D.E.M. is on scientific advisory boards at Oxford Nanopore Technologies (ONT) and Basis Genetics, is engaged in a research agreement with ONT, has received research and travel support from ONT and PacBio, and holds stock options in MyOme and Basis Genetics. All n-Lorem authors are compensated by n-Lorem. S.T.C., Chairman and CEO of n-Lorem receives no compensation. As a nonprofit n-Lorem does not provide stock options or RSUs.
Figures

Similar articles
-
Developing splice-switching oligonucleotides for urea cycle disorder using an integrated diagnostic and therapeutic platform.J Hepatol. 2025 Aug;83(2):411-425. doi: 10.1016/j.jhep.2025.02.007. Epub 2025 Feb 18. J Hepatol. 2025. PMID: 39978599
-
Deep learning analyses of splicing variants identify the link of PCP4 with amyotrophic lateral sclerosis.Brain. 2025 Jul 7;148(7):2331-2347. doi: 10.1093/brain/awaf025. Brain. 2025. PMID: 39852553
-
OpenASO: RNA Rescue-designing splice-modulating antisense oligonucleotides through community science.RNA. 2025 Jul 16;31(8):1091-1102. doi: 10.1261/rna.080288.124. RNA. 2025. PMID: 40425312
-
Characterization of cognitive impairment in adult polyglucosan body disease.J Neurol. 2022 Jun;269(6):2854-2861. doi: 10.1007/s00415-022-10960-z. Epub 2022 Jan 8. J Neurol. 2022. PMID: 34999962 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Bigio EH, Weiner MF, Bonte FJ et al. Familial dementia due to adult polyglucosan body disease. Clin Neuropathol. 1997; 16:55–62. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources

