sRNA-mediated crosstalk between cell wall stress and galactose metabolism in Staphylococcus aureus
- PMID: 40671522
- PMCID: PMC12266140
- DOI: 10.1093/nar/gkaf616
sRNA-mediated crosstalk between cell wall stress and galactose metabolism in Staphylococcus aureus
Abstract
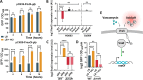
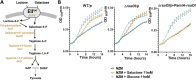
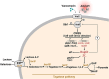
Staphylococcus aureus is an opportunistic pathogen responsible for a wide range of diseases in humans. During infections, this bacterium is exposed to various stresses that target its cell wall, such as oxidative or acid environments as well as various cell wall-acting antimicrobials. Staphylococcus aureus has effective regulatory systems for responding to environmental stresses, enabling the expression of factors necessary for its survival. Bacterial small RNAs (sRNAs) play a crucial role in this adaptation process. In this study, we show that RsaOI, an S. aureus sRNA, accumulates under acid stress conditions. This response is mediated via the two-component system VraSR, which is associated with the cell wall damage response. As a component of the VraSR regulon, RsaOI contributes to the survival of S. aureus under acid stress and affects its susceptibility to glycopeptide antibiotics. Our findings reveal that RsaOI targets the lacABCDFEG operon, which encodes components of tagatose pathway, a unique mechanism responsible for galactose metabolism in S. aureus. By antisense base pairing near the ribosome binding site of lacD, RsaOI inhibits the expression of this gene, encoding tagatose-6-phosphate aldolase. This regulation disrupts the tagatose pathway, impairing galactose utilization in S. aureus. These findings highlight the role of RsaOI in the mediation between cell wall stress responses and a specific metabolic pathway.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

