A computational dynamic model of combination treatment for type II inhibitors with asciminib
- PMID: 40671559
- PMCID: PMC12268125
- DOI: 10.1002/pro.70219
A computational dynamic model of combination treatment for type II inhibitors with asciminib
Abstract
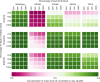
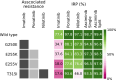
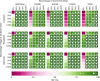
Despite continuous strides forward in drug development, resistance to treatment looms large in the battle against cancer as well as communicable diseases. Chronic myeloid leukemia (CML) is treated with targeted therapy and treatment is personalized when resistance arises. It has been extensively studied and is used as a model for targeted therapy. In this study, we examine combination treatments of type II Abl1 inhibitors and asciminib (an allosteric regulator) through a computational model at patient relevant concentrations. Due to the separate binding sites of type II inhibitors and asciminib, we propose their combination treatment as potentially robust to resistance. We find that the simultaneous cobinding of type II inhibitors and asciminib is high in synergetic combinations. As an aid to designing and comparing combination treatments, we put forward an equation that expands on the previously published effective ratio of IC50 (ERIC). Unlike usual comparisons of IC50 values, ERIC takes patient plasma concentrations into account. This study shows that the product of two ERIC values ( ) creates comparable approximations of the effectiveness of combination treatments with low levels of synergy or antagonism at different concentrations. Its simple formulation is done without experiments and requires less computation and input data than the current standard of ZIP values. As such, the new scheme is a useful complement to experiments that deal with synergy in drug use.
Keywords: asciminib; kinase inhibitor; leukemia; resistance mutations.
© 2025 The Author(s). Protein Science published by Wiley Periodicals LLC on behalf of The Protein Society.
Figures






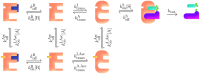

References
-
- Abumiya M, Takahashi N, Yoshioka T, Kameoka Y, Miura M. Evaluation of the plasma concentration of ponatinib in a chronic myeloid leukaemia patient with ponatinib intolerance. J Clin Pharm Ther. 2020;46(1):219–222. - PubMed
-
- Advani AS, Pendergast AM. Bcr–abl variants: biological and clinical aspects. Leuk Res. 2002;26(8):713–720. - PubMed
-
- Albe KR, Butler MH, Wright BE. Cellular concentrations of enzymes and their substrates. J Theor Biol. 1990;143(2):163–195. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

