This is a preprint.
Capturing Unanticipated Drug Toxicities Using an Ensemble Machine Learning Approach
- PMID: 40671799
- PMCID: PMC12265155
- DOI: 10.21203/rs.3.rs-6999821/v1
Capturing Unanticipated Drug Toxicities Using an Ensemble Machine Learning Approach
Abstract
Despite rigorous safety evaluations during development, numerous drugs have been withdrawn from the market due to serious toxicities. Here we investigate the features found in drugs with these unanticipated toxicities and apply a machine learning approach to predict if a drug is likely to be withdrawn due to intolerable side effects without the need for human trial data. Our best preforming classifier was an ensemble predictor trained on protein targets, protein structure features, chemical fingerprints, and chemical features that achieved 92% accuracy and 0.845 Matthews Correlation Coefficient with 10-fold holdout test set cross validation. Analysis of features predictive of unanticipated toxicity revealed both known factors such as inhibition of cytochrome P450 as well as yet uninvestigated factors including the inhibition of bile salt export pumps. This predictor and subsequent feature analysis pave the way for the larger role of computational methods in screening potential candidates during drug development.
Conflict of interest statement
COMPETING INTERESTS The authors have no conflicts of interest to declare. Additional Declarations: No competing interests reported.
Figures
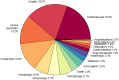
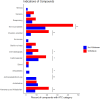
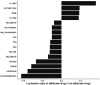

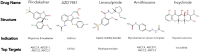
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

