ECM1 expression in chronic liver disease: Regulation by EGF/STAT1 and IFNγ/NRF2 signalling
- PMID: 40671832
- PMCID: PMC12260417
- DOI: 10.1016/j.jhepr.2025.101423
ECM1 expression in chronic liver disease: Regulation by EGF/STAT1 and IFNγ/NRF2 signalling
Abstract
Background & aims: The extracellular matrix protein 1 (ECM1) is essential for liver homeostasis by keeping latent transforming growth factor-beta quiescent. Upon hepatocyte damage, ECM1 is significantly downregulated, facilitating fibrosis and chronic liver disease (CLD) progression. We investigated the mechanism of ECM1 regulation in hepatocytes under pathophysiological conditions.
Methods: We used promoter analysis to predict Ecm1 transcriptional regulators and assessed the expression of Ecm1-related genes by single-cell RNA sequencing (scRNA-seq) and bulk RNA-seq. Functional assays were performed with AML12 cells, mouse and human primary hepatocytes, and liver tissue from mice and patients.
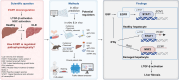
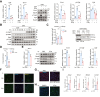
Results: In healthy hepatocytes, epidermal growth factor (EGF)/EGF receptor (EGFR) signalling sustains ECM1 expression through phosphorylating signal transducer and activator of transcription 1 (STAT1) at serine727 (S727), thus enhancing its binding to the ECM1 promoter and boosting gene transcription. This process is disrupted during liver inflammation by interferon gamma (IFNγ), which downregulates EGFR and inhibits EGF/EGFR/STAT1-mediated ECM1 promoter binding. Mechanistically, IFNγ-induced STAT1 phosphorylation at tyrosine701 (Y701) impairs the binding of p-STAT1 S727 to the ECM1 promoter. Additionally, IFNγ induces nuclear factor erythroid 2-related factor 2 (NRF2) nuclear translocation, which repressively binds to the promoter of ECM1, further reducing its expression. These findings are confirmed in several CLD mouse models (n = 2-6). Moreover, AAV8-ECM1 attenuates liver fibrosis and injury in Western diet-fed mice (n = 8-10), counteracting the effects of EGF signalling inhibition and IFNγ/NRF2 activation. In CLD patients (n = 22), ECM1 levels correlate positively with EGFR expression (p <0.0001) and negatively with IFNγ/NRF2 activation (p <0.0001).
Conclusions: EGF/STAT1 signalling promotes whereas IFNγ/NRF2 inhibits ECM1 expression in hepatocytes in health or disease, respectively. ECM1 has the potential to be developed as an antifibrotic agent, particularly in inflammation- or reactive oxygen species-driven CLD.
Impact and implications: This study reveals the regulatory mechanism of ECM1 in hepatocytes, demonstrating that EGF/STAT1 maintains ECM1 expression to prevent fibrosis, whereas IFNγ/NRF2 signalling inhibits ECM1 during chronic liver inflammation, thereby accelerating disease progression. These findings are important for researchers and clinicians to understand the pathogenesis of liver fibrosis, especially in CLD driven by inflammation or oxidative stress. Clinically, ECM1 levels correlate positively with EGFR expression and negatively with IFNγ/NRF2 activation, providing potential antifibrotic targets for CLD patients.
Keywords: Extracellular matrix; Growth factor; IFNγ; Liver fibrosis; TGF-β.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest that pertain to this work. Please refer to the accompanying ICMJE disclosure forms for further details.
Figures






References
-
- Mathieu E., Meheus L., Raymackers J., et al. Characterization of the osteogenic stromal cell line MN7: identification of secreted MN7 proteins using two-dimensional polyacrylamide gel electrophoresis, western blotting, and microsequencing. J Bone Miner Res. 1994;9:903–913. - PubMed
-
- Smits P., Poumay Y., Karperien M., et al. Differentiation-dependent alternative splicing and expression of the extracellular matrix protein 1 gene in human keratinocytes. J Invest Dermatol. 2000;114:718–724. - PubMed
-
- Sercu S., Lambeir A.M., Steenackers E., et al. ECM1 interacts with fibulin-3 and the beta 3 chain of laminin 332 through its serum albumin subdomain-like 2 domain. Matrix Biol. 2009;28:160–169. - PubMed
-
- Wang L., Yu J., Ni J., et al. Extracellular matrix protein 1 (ECM1) is over-expressed in malignant epithelial tumors. Cancer Lett. 2003;200:57–67. - PubMed
-
- Sercu S., Zhang L., Merregaert J. The extracellular matrix protein 1: its molecular interaction and implication in tumor progression. Cancer Invest. 2008;26:375–384. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

