The molecular sub-type and the development and validation of a prognosis prediction model based on endocytosis-related genes for hepatocellular carcinoma
- PMID: 40672080
- PMCID: PMC12260949
- DOI: 10.21037/jgo-2025-359
The molecular sub-type and the development and validation of a prognosis prediction model based on endocytosis-related genes for hepatocellular carcinoma
Abstract
Background: Despite the critical role of endocytosis-related genes in oncogenic processes, research exploring their potential for prognosticating hepatocellular carcinoma (HCC) remains limited. Establishing a connection between endocytosis and HCC is imperative. This study aimed to create a gene signature related to endocytosis to identify HCC subtypes and predict outcomes.
Methods: RNA sequencing and clinical data of 371 HCC patients were obtained from The Cancer Genome Atlas (TCGA)-HCC dataset. Subtypes of HCC were identified through endocytosis-associated genes through consistent clustering analysis, and prognosis was assessed using an endocytosis-associated HCC model. Construction and validation of a prognostic endocytosis-related risk scoring system were created for HCC.
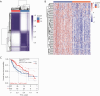
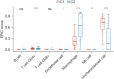
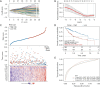
Results: A univariate Cox regression analysis was performed using the TCGA-HCC dataset, resulting in the identification of 4,354 genes significantly associated with patient prognosis. Subsequent Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of these genes identified several biologically relevant pathways, particularly those related to endocytosis, autophagy, and cell cycle regulation. Through the application of consensus clustering methods, patients with TCGA-HCC were stratified into two distinct subtypes based on a selection of 82 genes associated with endocytosis. Importantly, the overall survival rate for the high-risk subtype (C1) was significantly higher than that of the low-risk subtype (C2). KEGG analysis indicated that the upregulated genes in the high-risk C1 subtype were predominantly related to various pathways, including the p53 signaling pathway, proteoglycans in cancer, cell cycle regulation, interactions between the extracellular matrix and receptors, and cellular senescence. In contrast, in the comparison between the C1 and C2 HCC samples, the genes exhibiting downregulation were predominantly linked to metabolic pathways, including tyrosine metabolism and steroid hormone biosynthesis. Boxplots showed significant differences in immune cell populations, including CD4+ T lymphocytes, endothelial cells, natural killer cells, and macrophages. From a pool of 82 endocytosis-related genes, 14 genes were identified through least absolute shrinkage and selection operator and Cox regression, including CLTA, STAM, RAB10, DAB2, VPS45, AGAP3, ARPC4, VPS29, HSPA8, DNAJC6, PARD6B, ACTR3B, PSD4, and ARRB2. Based on these genetic markers, patients were stratified into low-risk and high-risk categories. The prognostic performance of the model was validated using receiver operating characteristic curve analysis, which produced area under the curve values of 0.807, 0.757, and 0.716 for 1-, 3-, and 5-year survival predictions, respectively. The model of endocytosis-related genes was validated by external International Cancer Genome Consortium (ICGC)-HCC datasets.
Conclusions: Genes linked to endocytosis strongly correlate with tumor classification in patients with HCC. The related expression profiles may be valuable for predicting HCC prognosis and informing diagnosis and treatment.
Keywords: Hepatocellular carcinoma (HCC); biomarkers; endocytosis; prognostic model.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-359/coif). The authors have no conflicts of interest to declare.
Figures



Similar articles
-
PANoptosis-associated genes exhibit significant potential in the diagnosis of hepatocellular carcinoma.J Gastrointest Oncol. 2025 Jun 30;16(3):1105-1114. doi: 10.21037/jgo-2025-356. Epub 2025 Jun 27. J Gastrointest Oncol. 2025. PMID: 40672091 Free PMC article.
-
A novel prognostic model based on immunogenic cell death-related genes for improved risk stratification in hepatocellular carcinoma patients.J Cancer Res Clin Oncol. 2023 Sep;149(12):10255-10267. doi: 10.1007/s00432-023-04950-5. Epub 2023 Jun 3. J Cancer Res Clin Oncol. 2023. PMID: 37269346 Free PMC article.
-
Anoikis-related lncRNA signature predicts prognosis and is associated with immune infiltration in hepatocellular carcinoma.Anticancer Drugs. 2024 Jun 1;35(5):466-480. doi: 10.1097/CAD.0000000000001589. Epub 2024 Mar 11. Anticancer Drugs. 2024. PMID: 38507233
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
