Identification and validation of a ferroptosis-related long non-coding RNA signature as a prognostic biomarker for hepatocellular carcinoma
- PMID: 40672083
- PMCID: PMC12261030
- DOI: 10.21037/jgo-2025-360
Identification and validation of a ferroptosis-related long non-coding RNA signature as a prognostic biomarker for hepatocellular carcinoma
Abstract
Background: Ferroptosis may play a central role in the development of hepatocellular carcinoma (HCC). Ferroptosis-related long non-coding RNAs (FRlncRNAs) show prognostic value in HCC through ferroptosis regulation and significant survival correlation. This study aimed to identify a prognostic FRlncRNA signature and explored its association with tumor immunity and oncogenic pathways.
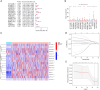
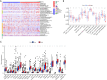
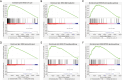
Methods: The transcriptome sequencing information and matched clinical data of 365 HCC patients was obtained from The Cancer Genome Atlas (TCGA) database. Patients were randomly divided into training and testing groups. A prognostic FRlncRNA signature was established in the test set via a correlation analysis, univariate Cox regression analysis, and LLeast Absolute Shrinkage and Selection Operator (LASSO) regression analysis. We evaluated the signature's prognostic significance through clinical correlation analysis and developed a predictive nomogram for HCC outcomes. Immune function and checkpoint analyses were conducted to explore the association between the signature and HCC-related immunity. Additionally, a Gene Set Enrichment Analysis (GSEA) was conducted to detect the enriched pathways. Among genes identified, the oncogenic function of LINC01063 was studied both in vitro and in vivo.
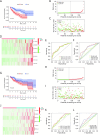
Results: In the training set, we established a prognostic signature comprising seven FRlncRNAs and classified patients into low-risk (LR) and high-risk (HR) groups with different prognosis. Time-dependent receiver operating characteristic (ROC) analysis yielded area under the ROC curve (AUC) values of 0.745, 0.745, and 0.719 for 1-, 2-, and 3-year overall survival (OS). The prognostic impact of risk-groups was verified in the testing set. The HR patients exhibited greater infiltration of immune cells and elevated expression levels of immune checkpoint genes. Significant differences in the cytolytic activity and Type II interferon response between the LR and HR groups were found. Several signaling pathways were enriched in the HR group, indicating that the signature was closely associated with HCC development. Finally, among the seven FRlncRNAs in the signature, LINC01063 was validated as an oncogene. In vitro, the knockdown of LINC01063 inhibited cell proliferation, disrupted colony formation ability, and reduced the migration and invasion capacities of HCC cells, and in vivo, nude BALB/c mice injected with the LINC01063-knockdown HCC cells exhibited reduced tumor growth compared to the controls.
Conclusions: A signature of seven FRlncRNAs predicted outcome and correlated with immunity and activated oncogene pathways suggesting that the signature could be a predictor of efficacy of immunotherapy. LINC01063 was validated as a new oncogene in HCC. Our findings provide novel insights for prognostic assessment and precision therapy in HCC.
Keywords: Hepatocellular carcinoma (HCC); LINC01063 oncogene; ferroptosis; long non-coding RNA (lncRNA); prognostic signature.
Copyright © 2025 AME Publishing Company. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://jgo.amegroups.com/article/view/10.21037/jgo-2025-360/coif). M.L. reports unrestricted research funding from Scandion Oncology A/S, Copenhagen, Denmark, and is advisory board member of Alivia AB, Stockholm, Sweden. The other authors have no conflicts of interest to declare.
Figures



References
-
- Huang E, Ma T, Zhou J, et al. The development and validation of a novel senescence-related long-chain non-coding RNA (lncRNA) signature that predicts prognosis and the tumor microenvironment of patients with hepatocellular carcinoma. Ann Transl Med 2022;10:766. 10.21037/atm-22-3348 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
