This is a preprint.
Immunosuppressants Rewire the Gut Microbiome-Alloimmune Axis Through Time-Dependent and Tissue-Specific Mechanisms
- PMID: 40672154
- PMCID: PMC12265612
- DOI: 10.1101/2025.01.02.631100
Immunosuppressants Rewire the Gut Microbiome-Alloimmune Axis Through Time-Dependent and Tissue-Specific Mechanisms
Abstract
Background: Lifelong immunosuppressive therapy is required to prevent allograft rejection in organ transplantation. Current immunosuppressants effectively suppress adaptive and innate immune responses, but their broad, antigen-non-specific effects often result in severe off-target complications. It remains a significant unmet medical need in transplant medicine.
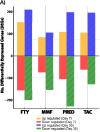
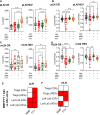
Results: In this study we investigated immunosuppressant effects of four major immunosuppressant classes, including tacrolimus, prednisone, mycophenolate mofetil (MMF), and fingolimod (FTY), on the gut microbiome, metabolic pathways, lymphoid architecture and lymphocyte trafficking after up to 30-day chronic exposure. Despite their distinct mechanisms of action and not designed to target the gut, all immunosuppressive drugs induced profound and time-dependent alterations in both intestine gene expression and gut microbiome composition. Progressive alterations from moderate early, drug-specific changes to a strikingly convergent microbial dysbiosis, marked by significant expansion of pathobionts of Muribaculaceae, occurred across all drug classes. Concurrently, all drugs uniformly induced significant suppression of mucosal immunity including B cell, immunoglobulin, and antigen recognition. Time-dependent changes in lymph node (LN) reorganization and cellular composition were also observed, marked by a progressive shift toward pro-inflammatory phenotypes in gut-draining mesenteric LNs and a gradual loss of tolerogenic architecture in peripheral LNs. Drug-specific metabolic alterations and distinct phases of intestinal transcriptional responses were also characterized. Notably, MMF and FTY demonstrated the most robust immunomodulatory properties, and were able to suppress alloantigen-induced inflammation through mediating regulatory T cell distribution and LN remodeling.
Conclusions: Together, these findings highlight the underappreciated complexity and temporal dynamics immunosuppressants effects, particularly their impact on the gut and compartmentalized regulation of alloimmune in lymphoid tissues. Understanding these relationships offers new opportunities for refining immunosuppressive strategies to reduce treatment-related off-target complications and improve long-term organ transplant outcomes.
Keywords: gut dysbiosis; immunology; immunosuppressants; metabolome; microbiome.
Conflict of interest statement
Competing interests No potential financial and non-financial conflict of interest was reported by the authors.
Figures







Similar articles
-
Clinical and cost-effectiveness of newer immunosuppressive regimens in renal transplantation: a systematic review and modelling study.Health Technol Assess. 2005 May;9(21):1-179, iii-iv. doi: 10.3310/hta9210. Health Technol Assess. 2005. PMID: 15899149
-
Synbiotics, prebiotics and probiotics for solid organ transplant recipients.Cochrane Database Syst Rev. 2022 Sep 20;9(9):CD014804. doi: 10.1002/14651858.CD014804.pub2. Cochrane Database Syst Rev. 2022. PMID: 36126902 Free PMC article.
-
A systematic review and economic model of the clinical and cost-effectiveness of immunosuppressive therapy for renal transplantation in children.Health Technol Assess. 2006 Dec;10(49):iii-iv, ix-xi, 1-157. doi: 10.3310/hta10490. Health Technol Assess. 2006. PMID: 17134597
-
T-bet expressing Tr1 cells driven by dietary signals dominate the small intestinal immune landscape.bioRxiv [Preprint]. 2025 Jul 4:2025.06.30.662190. doi: 10.1101/2025.06.30.662190. bioRxiv. 2025. PMID: 40747421 Free PMC article. Preprint.
-
Immunosuppressants for the prophylaxis of corneal graft rejection after penetrating keratoplasty.Cochrane Database Syst Rev. 2015 Aug 27;2015(8):CD007603. doi: 10.1002/14651858.CD007603.pub2. Cochrane Database Syst Rev. 2015. PMID: 26313245 Free PMC article.
References
-
- Schmitz V., Klawitter J., Bendrick-Peart J., Schoening W., Puhl G., Haschke M., Klawitter J., Consoer J., Rivard C.J., Chan L., et al. (2009). Metabolic profiles in urine reflect nephrotoxicity of sirolimus and cyclosporine following rat kidney transplantation. Nephron Exp Nephrol 111, e80–91. 10.1159/000209208. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
