This is a preprint.
Identification and characterization of functional and trans-dominant negative HERV-K (HML-2) Rec proteins encoded in the human genome
- PMID: 40672174
- PMCID: PMC12265724
- DOI: 10.1101/2025.07.07.663474
Identification and characterization of functional and trans-dominant negative HERV-K (HML-2) Rec proteins encoded in the human genome
Abstract
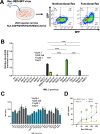
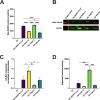
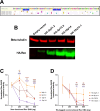
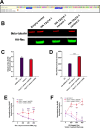
Human Endogenous Retroviruses K (HERV-K) of the HML-2 subgroup are the most recently integrated and biologically active retroviral elements within the human genome. The HERV-K Rec protein, a functional homolog of HIV Rev and HTLV Rex, is necessary for the nuclear export of viral mRNAs with retained introns. However, the diversity of Rec proteins encoded in the human genome and their functional capacities have remained largely unexplored. We identified full-length Rec protein sequences in the human genome and selected 23 variants for functional characterization. Using a dual-color fluorescent reporter and a complementary ELISA assay, we found that Rec proteins from only 7 genomic loci were functional. In addition, a subset of the non-functional Rec proteins exerted trans-dominant negative effects. Detailed mutational analysis of the most potent inhibitory variant, encoded by the HERV-K provirus 12q14.1, displayed only 2 amino acid changes (H2N and del34E) relative to the prototypical functional Rec protein encoded by the reconstructed consensus HERV-K (HERV-K Con). Insertion of a glutamic acid at position 34 restored the functional activity, while the substitution of histidine with asparagine at position 2 did not. Our results unveil an unexpected complexity in HERV-K (HML-2) post-transcriptional regulation, with Rec variants displaying a spectrum of activities ranging from robust nuclear export of RNA with retained introns to trans-dominant negative inhibition of Rec function. These findings expand our understanding of the regulatory landscape governing HERV-K (HML-2) expression and suggest mechanisms by which different HERV-K Rec proteins may influence host cell biology and pathology, including oncogenesis.
Figures

Similar articles
-
Investigating the expression of HERV-K env, np9, gag, and rec in bladder cancers.Infect Agent Cancer. 2025 Jun 21;20(1):41. doi: 10.1186/s13027-025-00665-z. Infect Agent Cancer. 2025. PMID: 40544283 Free PMC article.
-
The expression analysis of human endogenous retrovirus-K Env, Np9, and Rec transcripts in cervical cancer.J Med Virol. 2024 Mar;96(3):e29501. doi: 10.1002/jmv.29501. J Med Virol. 2024. PMID: 38445563
-
An integrated approach for the accurate detection of HERV-K HML-2 transcription and protein synthesis.Nucleic Acids Res. 2025 Jan 11;53(2):gkaf011. doi: 10.1093/nar/gkaf011. Nucleic Acids Res. 2025. PMID: 39831303 Free PMC article.
-
Magnetic resonance perfusion for differentiating low-grade from high-grade gliomas at first presentation.Cochrane Database Syst Rev. 2018 Jan 22;1(1):CD011551. doi: 10.1002/14651858.CD011551.pub2. Cochrane Database Syst Rev. 2018. PMID: 29357120 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
-
- Bannert N, Kurth R. 2006. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet 7:149–73. - PubMed
-
- Katzourakis A, Rambaut A, Pybus OG. 2005. The evolutionary dynamics of endogenous retroviruses. Trends Microbiol 13:463–8. - PubMed
-
- Stoye JP. 2012. Studies of endogenous retroviruses reveal a continuing evolutionary saga. Nat Rev Microbiol 10:395–406. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
