This is a preprint.
Multiple-testing corrections in case-control studies using identity-by-descent segments
- PMID: 40672175
- PMCID: PMC12265659
- DOI: 10.1101/2025.07.03.663057
Multiple-testing corrections in case-control studies using identity-by-descent segments
Abstract
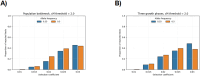
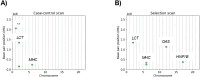
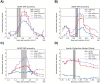
Identity-by-descent (IBD) mapping provides complementary signals to genome-wide association studies (GWAS) when multiple causal haplotypes or variants are present, but not directly tested. However, failing to correct for multiple testing in case-control studies using IBD segments can lead to false discoveries. We propose the difference between case-case and control-control IBD rates as an IBD mapping statistic. For our hypothesis test, we use a computationally efficient approach from the stochastic processes literature to derive genome-wide significance levels that control the family-wise error rate (FWER). Whole genome simulations indicate that our method conservatively controls the FWER. Because positive selection can lead to false discoveries, we pair our IBD mapping approach with a selection scan so that one can contrast results for evidence of confounding due to recent sweeps or other mechanisms, like population structure, that increase IBD sharing. We developed automated and reproducible workflows to phase haplotypes, call local ancestry probabilities, and perform the IBD mapping scan, the former two tasks being important preprocessing steps for haplotype analyses. We applied our methods to search for Alzheimer's disease (AD) risk loci in the Alzheimer's Disease Sequencing Project (ADSP) genome data. We identified six genome-wide significant signals of AD risk among samples genetically similar to African and European reference populations and self-identified Amish samples. Variants in the six potential risk loci we detected have previously been associated with AD, dementia, and memory decline. Three genes at two potential risk loci have already been nominated as therapeutic targets. Overall, our scalable approach makes further use of large consortia resources, which are expensive to collect but provide insights into disease mechanisms.
Keywords: Alzheimer’s disease; binary traits; haplotypes; identity by descent; mean-reverting processes; multiple testing.
Conflict of interest statement
Declaration of interests T.A.T. is a current employee of Regeneron Genetics Center and stockholder of Regeneron Pharmaceuticals. The other authors declare no competing interests.
Figures
Similar articles
-
Multiple-testing corrections in selection scans using identity-by-descent segments.bioRxiv [Preprint]. 2025 Jan 29:2025.01.29.635528. doi: 10.1101/2025.01.29.635528. bioRxiv. 2025. PMID: 39975073 Free PMC article. Preprint.
-
Identity by descent and local ancestry mapping of HCV spontaneous clearance in populations of diverse ancestries.BMC Genomics. 2025 Jul 12;26(1):661. doi: 10.1186/s12864-024-11076-6. BMC Genomics. 2025. PMID: 40652174 Free PMC article.
-
Regional cerebral blood flow single photon emission computed tomography for detection of Frontotemporal dementia in people with suspected dementia.Cochrane Database Syst Rev. 2015 Jun 23;2015(6):CD010896. doi: 10.1002/14651858.CD010896.pub2. Cochrane Database Syst Rev. 2015. PMID: 26102272 Free PMC article.
-
Electronic cigarettes for smoking cessation and reduction.Cochrane Database Syst Rev. 2014;(12):CD010216. doi: 10.1002/14651858.CD010216.pub2. Epub 2014 Dec 17. Cochrane Database Syst Rev. 2014. PMID: 25515689
-
Plasma and cerebrospinal fluid amyloid beta for the diagnosis of Alzheimer's disease dementia and other dementias in people with mild cognitive impairment (MCI).Cochrane Database Syst Rev. 2014 Jun 10;2014(6):CD008782. doi: 10.1002/14651858.CD008782.pub4. Cochrane Database Syst Rev. 2014. PMID: 24913723 Free PMC article.
References
-
- Tam V., Patel N., Turcotte M., Bossé Y., Paré G., Meyre D., Benefits and limitations of genome-wide association studies, Nat. Rev. Genet. 20 (2019) 467–484. - PubMed
-
- Uffelmann E., Huang Q. Q., Munung N. S., de Vries J., Okada Y., Martin A. R., Martin H. C., Lappalainen T., Posthuma D., Genome-wide association studies, Nat. Rev. Methods Primers 1 (2021) 1–21.
-
- Albrechtsen A., Sand Korneliussen T., Moltke I., van Overseem Hansen T., Nielsen F. C., Nielsen R., Relatedness mapping and tracts of relatedness for genome-wide data in the presence of linkage disequilibrium, Genetic Epidemiology 33 (2009) 266–274. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
