This is a preprint.
Sex chromosome identification and genome curation from a single individual with SCINKD
- PMID: 40672204
- PMCID: PMC12265721
- DOI: 10.1101/2025.07.07.660342
Sex chromosome identification and genome curation from a single individual with SCINKD
Abstract
In most animal species, the sex determining pathway is typically initiated by the presence/absence of a primary genetic cue at a critical point during development. This primary genetic cue is often located on a single locus-referred to as sex chromosomes-and can be limited to females (in a ZZ/ZW system) or males (in an XX/XY system). One trademark of sex chromosomes is a restriction or cessation of recombination surrounding the sex-limited region (to prevent its inheritance in the homogametic sex). This may lead to-through a variety of mechanisms-higher amounts of genetic divergence within this region, i.e. between the X/Z and Y/W chromosomes, especially when compared to their autosomal counterparts. Recent advances in genome sequencing and computation have brought with them the ability to resolve haplotypes within a diploid individual, permitting assembly of previously challenging genomic regions like sex chromosomes. Leveraging these advances, we identified replicable diagnostic characteristics between typical autosomes and sex chromosomes (within a single genome assembly). Under this framework, we can use this information to identify putative sex chromosome linkage groups across divergent vertebrate taxa and simultaneously curate misassembled regions on autosomes. Here, we present this conceptual framework and associated tool for identifying candidate sex chromosome linkage groups from a single, diploid individual dubbed Sex Chromosome Identification by Negating Kmer Densities, or SCINKD.
Conflict of interest statement
Conflict of interest statement: None declared.
Figures
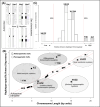






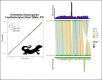


References
-
- Antipov D., Rautiainen M., Nurk S., Walenz B. P., Solar S. J., Phillippy A. M., & Koren S. (2024). Verkko2: Integrating proximity ligation data with long-read De Bruijn graphs for efficient telomere-to-telomere genome assembly, phasing, and scaffolding. In bioRxiv. 10.1101/2024.12.20.629807 - DOI - PMC - PubMed
-
- Bergero R., & Charlesworth D. (2009). The evolution of restricted recombination in sex chromosomes. Trends in Ecology & Evolution, 24(2), 94–102. - PubMed
Publication types
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
