This is a preprint.
PRMT5 activity sustains histone production to maintain genome integrity
- PMID: 40672221
- PMCID: PMC12265569
- DOI: 10.1101/2025.07.03.663002
PRMT5 activity sustains histone production to maintain genome integrity
Abstract
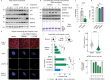
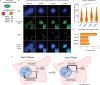
Histone proteins package DNA into nucleosomes, forming chromatin and thereby safeguarding genome integrity. Proper histone expression is essential for cell proliferation and chromatin organization, yet the upstream regulators of histone supply remain incompletely understood. PRMT5-a cell essential type II protein arginine methyltransferase frequently overexpressed in cancer-catalyzes symmetric dimethylation of arginine residues. Using time-resolved nascent transcriptional profiling, quantitative proteomics, and imaging, we show that PRMT5 activity is required to sustain histone transcription and histone protein synthesis during S phase. PRMT5 inhibition or knockdown leads to rapid histone mRNA depletion, loss of histone proteins, and accumulation of replication-associated nuclear abnormalities. We further show that soluble histone H4 accumulates at histone locus bodies (HLBs) upon PRMT5 inhibition, and that PRMT5-substrate H4 Arginine 3 mutants localize more robustly to HLBs than do wildtype H4. These findings support a model in which PRMT5-mediated methylation of histone H4 regulates histone transcription. Our findings establish PRMT5 as a central coordinator of histone homeostasis and provide a mechanistic rationale for its essential role in proliferating cells.
Keywords: Arginine Methylation; Genome Integrity; H1; H4R3C; H4R3me2s; Histone H4; Histone Transcription; Methyltransferase; PRMT5; Post-Translational Modifications; methylarginine; micronuclei; oncohistone; symmetric dimethylation.
Conflict of interest statement
Conflict of Interest The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. J.B. was an employee of Arpeggio Bio, and J.A. is an employee and founder of Arpeggio Bio, which was contracted to undertake the PRO-seq experiments described in this paper.
Figures





Similar articles
-
Cardiac-specific overexpression of PRMT5 exacerbates pressure overload-induced hypertrophy and heart failure.J Biomed Sci. 2025 Jul 6;32(1):61. doi: 10.1186/s12929-025-01162-6. J Biomed Sci. 2025. PMID: 40619438 Free PMC article.
-
The Histone Methyltransferase PRMT5 Mediates the Epigenetic Modification to Modulate High Temperatures and Tea Quality in Tea Plants (Camellia sinensis).Plant Cell Environ. 2025 Aug;48(8):5973-5992. doi: 10.1111/pce.15567. Epub 2025 Apr 23. Plant Cell Environ. 2025. PMID: 40269587
-
Core histones govern echinocandin susceptibility in Candida glabrata.Microbiol Spectr. 2025 Jun 3;13(6):e0239924. doi: 10.1128/spectrum.02399-24. Epub 2025 Apr 30. Microbiol Spectr. 2025. PMID: 40304478 Free PMC article.
-
Dismantling the epigenetic alliance: Emerging strategies to disrupt the PRMT5:MEP50 complex for cancer therapy.Eur J Med Chem. 2025 Oct 15;296:117870. doi: 10.1016/j.ejmech.2025.117870. Epub 2025 Jun 14. Eur J Med Chem. 2025. PMID: 40532498 Review.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
