This is a preprint.
The mevalonate pathway of isoprenoid biosynthesis supports metabolic flexibility in Mycobacterium marinum
- PMID: 40672248
- PMCID: PMC12265665
- DOI: 10.1101/2025.07.11.664281
The mevalonate pathway of isoprenoid biosynthesis supports metabolic flexibility in Mycobacterium marinum
Abstract
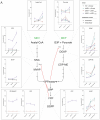
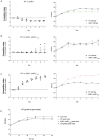
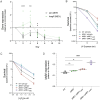
Isoprenoids are a diverse class of natural products that are essential in all domains of life. Most bacteria synthesize isoprenoids through either the methylerythritol phosphate (MEP) pathway or the mevalonate (MEV) pathway, while a small subset encodes both pathways, including the pathogen Mycobacterium marinum (Mm). It is unclear whether the MEV pathway is functional in Mm, or why Mm encodes seemingly redundant metabolic pathways. Here we show that the MEP pathway is essential in Mm while the MEV pathway is dispensable in culture, with the ΔMEV mutant having no growth defect in axenic culture but a competitive growth defect compared to WT Mm. We found that the MEV pathway does not play a role in ex vivo or in vivo infection but does play a role in survival of peroxide stress. Metabolite profiling revealed that modulation of the MEV pathway causes compensatory changes in the concentration of MEP intermediates DOXP and CDP-ME, suggesting that the MEV pathway is functional and that the pathways interact at the metabolic level. Finally, the MEV pathway is upregulated early in the shift down to hypoxia, suggesting that it may provide metabolic flexibility to this bacterium. Interestingly, we found that our complemented strains, which vary in copy number of the polyprenyl synthetase idsB2, responded differently to peroxide and UV stresses, suggesting a role for this gene as a determinant of downstream prenyl phosphate metabolism. Together, these findings suggest that MEV may serve as an anaplerotic pathway to make isoprenoids under stress conditions.
Figures




Similar articles
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Systemic Inflammatory Response Syndrome.2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jun 20. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31613449 Free Books & Documents.
-
Interventions for managing oral submucous fibrosis.Cochrane Database Syst Rev. 2024 Feb 28;2(2):CD007156. doi: 10.1002/14651858.CD007156.pub3. Cochrane Database Syst Rev. 2024. PMID: 38415846 Free PMC article.
-
Orthodontic treatment for crowded teeth in children.Cochrane Database Syst Rev. 2021 Dec 31;12(12):CD003453. doi: 10.1002/14651858.CD003453.pub2. Cochrane Database Syst Rev. 2021. PMID: 34970995 Free PMC article.
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
References
-
- Sacchettini JC, Poulter CD. 1997. Creating Isoprenoid Diversity. Science 277:1788–1789. - PubMed
-
- Avalos M, Garbeva P, Vader L, van Wezel GP, Dickschat JS, Ulanova D. 2021. Biosynthesis, evolution and ecology of microbial terpenoids. Nat Prod Rep 39:249–272. - PubMed
-
- Eberl M, Hintz M, Reichenberg A, Kollas A-K, Wiesner J, Jomaa H. 2003. Microbial isoprenoid biosynthesis and human γδ T cell activation. Febs Lett 544:4–10. - PubMed
-
- Heuston S, Begley M, Gahan CGM, Hill C. 2012. Isoprenoid biosynthesis in bacterial pathogens. Microbiology+ 158:1389–1401. - PubMed
