This is a preprint.
PARP1 promotes replication-independent DNA double-strand break formation after acute DNA-methylation damage
- PMID: 40672314
- PMCID: PMC12265532
- DOI: 10.1101/2025.07.10.663928
PARP1 promotes replication-independent DNA double-strand break formation after acute DNA-methylation damage
Abstract
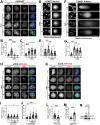
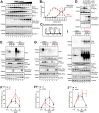
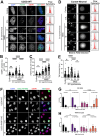
Poly-ADP-Ribose Polymerase 1 (PARP1) is a potent regulator of DNA damage response signaling through the recruitment of DNA damage repair proteins to damage sites, and its catalytic function of converting Nicotinamide adenine dinucleotide (NAD+) into poly-ADP-ribose (PAR) which covalently modifies hundreds of protein substrates in a process known as PARylation. However, PARP1's role in the recognition, processing, and intracellular signaling downstream of DNA damage in cells remains incompletely understood, especially in a replication-independent context. Here, we show that cells exposed to high doses of the methylating agent Methyl Methanesulfonate (MMS) generate DNA double-strand breaks (DSBs) in a base excision repair (BER)-dependent and DNA replication-independent manner. The capacity of cells to generate DSBs after MMS exposure relies heavily on intracellular NAD+ availability and PARP1's catalytic production of PAR. In our experimental system, we show that acute MMS exposure causes NAD+ exhaustion in a PARP1-dependent manner, which results in a temporal-dependent loss of downstream PARP1 activity. This functional loss of PARP1 signaling in later timepoints leads to the loss of BER-dependent single-strand break (SSB)-to-DSB conversion, as well as silencing of ATR-Chk1 signaling in both cycling and non-cycling cells, demonstrating a novel PARP1-dependent regulatory mechanism for both ATR-Chk1 signaling and BER-associated processes following methylation challenge. Additionally, we provide experimental evidence supporting the role of PARP1 and NAD+ in promoting the exonuclease-mediated SSB-to-DSB conversion. These findings support a previously uncharacterized mechanism of PARP1-mediated replication-independent DSB generation and provide insight into checkpoint signaling by integrating DDR with PARP1's consumption of NAD+ and production of PAR.
Conflict of interest statement
DECLARATION OF INTERESTS S.Y. and A.M.M. report a pending patent application related to this manuscript titled "Modulation of DNA double-strand break formation and repair by Poly-ADP-Ribose Polymerase 1, ADP-Ribosylation, and/or NAD+ availability to treat cancer" (PCT#63/783,282). Other authors declare that they have no competing interests.
Figures


Similar articles
-
Pold4 subunit of replicative polymerase δ promotes fork slowing at broken templates.DNA Repair (Amst). 2024 Jul;139:103688. doi: 10.1016/j.dnarep.2024.103688. Epub 2024 Apr 24. DNA Repair (Amst). 2024. PMID: 38678695
-
Molecular insights into PARP1 activation: structural dynamics of DNA, NAD+, and zinc‑mediated allosteric regulation.J Biomol Struct Dyn. 2025 Aug 26:1-27. doi: 10.1080/07391102.2025.2551909. Online ahead of print. J Biomol Struct Dyn. 2025. PMID: 40856521
-
Oncometabolite 2-hydroxyglutarate suppresses basal protein levels of DNA polymerase beta that enhances alkylating agent and PARG inhibition induced cytotoxicity.DNA Repair (Amst). 2024 Aug;140:103700. doi: 10.1016/j.dnarep.2024.103700. Epub 2024 Jun 4. DNA Repair (Amst). 2024. PMID: 38897003 Free PMC article.
-
NAD+-mediated regulation of mammalian base excision repair.DNA Repair (Amst). 2020 Sep;93:102930. doi: 10.1016/j.dnarep.2020.102930. DNA Repair (Amst). 2020. PMID: 33087267 Free PMC article. Review.
-
Joining of DNA breaks- interplay between DNA ligases and poly (ADP-ribose) polymerases.DNA Repair (Amst). 2025 May;149:103843. doi: 10.1016/j.dnarep.2025.103843. Epub 2025 May 2. DNA Repair (Amst). 2025. PMID: 40347914 Review.
References
-
- Brink A., Schulz B., Stopper H., and Lutz W.K. (2007). Biological significance of DNA adducts investigated by simultaneous analysis of different endpoints of genotoxicity in L5178Y mouse lymphoma cells treated with methyl methanesulfonate. Mutat Res 625, 94–101. 10.1016/j.mrfmmm.2007.05.007. - DOI - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
