This is a preprint.
The Global Landscape of Genetic Variation in Parkinson's disease: Multi-Ancestry Insights into Established Disease Genes and their Translational Relevance
- PMID: 40672482
- PMCID: PMC12265751
- DOI: 10.1101/2025.07.08.25330815
The Global Landscape of Genetic Variation in Parkinson's disease: Multi-Ancestry Insights into Established Disease Genes and their Translational Relevance
Abstract
Background: The genetic architecture of Parkinson's disease (PD) varies considerably across ancestries, yet most genetic studies have focused on individuals of European descent, limiting our insights into the genetic architecture of PD at a global scale.
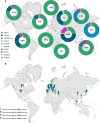
Methods: We conducted a large-scale, multi-ancestry investigation of causal and risk variants in PD-related genes. Using genetic datasets from the Global Parkinson's Genetics Program, we analyzed sequencing and genotyping data from 69,881 individuals, including 41,139 affected and 28,742 unaffected, from eleven different ancestries, including ~30% of individuals from non-European ancestries.
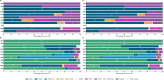
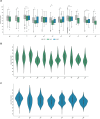
Findings: Our findings revealed shared and ancestry-specific patterns in the prevalence and spectrum of PD-associated variants. Overall, ~2% of affected individuals carried a causative variant, with substantial variations across ancestries ranging from <0·5% in African, African-admixed, and Central Asian to >10% in Middle Eastern and Ashkenazi Jewish ancestries. Including disease-associated GBA1 and LRRK2 risk variants raised the yield to ~12.5%, largely driven by GBA1, except in East Asians, where LRRK2 risk variants dominated. GBA1 variants were most frequent globally, albeit with substantial differences in frequencies and variant spectra. While GBA1 variants were identified across all ancestries, frequencies ranged from 3·4% in Middle Eastern to 51·7% in African ancestry. Similarly, LRRK2 variants showed ancestry-specific enrichment, with G2019S most frequently seen in Middle Eastern and Ashkenazi Jewish, and risk variants predominating in East Asians. However, clinical trials targeting proteins encoded by these genes are primarily based in Europe and North America.
Interpretation: This large-scale, multi-ancestry assessment offers crucial insights into the population-specific genetic architecture of PD. It underscores the critical need for increased diversity in PD genetic research to improve diagnostic accuracy, enhance our understanding of disease mechanisms across populations, and ensure the equitable development and application of emerging precision therapies.
Keywords: GBA1; Genetics; LRRK2; Multi-ancestry; PD; PRKN; SNCA; causal variants; precision medicine; risk variants.
Conflict of interest statement
LML, SoB, MLD, PH, SJ, SUR, LS, CS, SS, EJS, AHT, ZT, NZ, KL, and SBC declare no relevant conflict of interests. LML received faculty honoraria from the Movement Disorder Society in the past 12 months, unrelated to this manuscript. ZHF is supported by the Aligning Science Across Parkinson’s (ASAP) Global Parkinson’s Genetics Program (GP2) and receives GP2 salary support from The Michael J. Fox Foundation for Parkinson’s Research. MBM, NK, ShB, HI, LJ, MJK, HLL, KSL, DV, and MAN’s participation in this project was part of a competitive contract awarded to DataTecnica LLC by the National Institutes of Health to support open science research. MAN also currently serves on the scientific advisory board for Character Bio Inc plus is a scientific founder at Neuron23 Inc and owns stock. KAB and MT are supported by the Aligning Science Across Parkinson’s (ASAP) Global Parkinson’s Genetics Program (GP2). PH serves as an advisor to Alector Inc., the Global Parkinson’s Genetics Consortium (Michael J. Fox Foundation), LSP Advisory B.V., and Neuro.VC. HH and RK received essential funding from The Wellcome Trust, The MRC, The MSA Trust, The National Institute for Health Research University College London Hospitals Biomedical Research Centre NIHR-BRC), The Michael J Fox Foundation (MJFF), The Fidelity Trust, Rosetrees Trust, The Dolby Family fund, Alzheimer's Research UK (ARUK), MSA Coalition, The Guarantors of Brain, Cerebral Palsy Alliance, FARA, EAN and the NIH NeuroBioBank, Queen Square BrainBank, and The MRC Brainbank Network. JJ was supported by the Michael J. Fox Foundation (Data Community Innovators Program). KRK is supported by the Ainsworth 4 Foundation and the Medical Research Future Fund. SYL received consultancies and grants from the Michael J. Fox Foundation for Parkinson’s research (MJFF) and the Aligning Science Across Parkinson’s (ASAP) Global Parkinson’s Genetics Program (GP2). Unrelated to this manuscript, he received honoraria for participating as a Member of the Neurotorium Editorial Board and honoraria for lecturing/teaching from the International Parkinson and Movement Disorder Society (MDS) and Medtronic. He also received stipends from the MDS as Chair of the Asian-Oceanian Section, and npj PD as Associate Editor. NEM receives NIH funding (1K08NS131581). He is supported by the Aligning Science Across Parkinson’s (ASAP) Global Parkinson’s Genetics Program (GP2) and is member of the steering committee of the PD GENEration study for which he receives an honorarium from the Parkinson’s Foundation. WMYM is the founding member and Chair of the AfrAbia Parkinson’s Disease Genomic Consortium (AfrAbia PD-GC), which is supported by funding from the Global Parkinson’s Genetics Program (GP2). AJN reports grants from Parkinson's UK, Barts Charity, Cure Parkinson’s, National Institute for Health and Care Research, Innovate UK, the Medical College of Saint Bartholomew’s Hospital Trust, Alchemab, Aligning Science Across Parkinson’s Global Parkinson’s Genetics Program (ASAP-GP2) and the Michael J Fox Foundation and consultancy and personal fees from AstraZeneca, AbbVie, Profile, Bial, Charco Neurotech, Alchemab, Sosei Heptares, Umedeor and Britannia. He is an Associate Editor for the Journal of Parkinson’s Disease. RO has received travel grants from the Movement Disorder Society in the past 12 months, unrelated to this manuscript, and received research grants and travel support to attend annual GP2 meetings from the Aligning Science Across Parkinson’s (ASAP) Global Parkinson’s Genetics Program (GP2). NUO reports funding from MJFF and UK NIHR institutional grant funding, both for PD research. AHT receives support from the Michael J Fox Foundation and the Global Parkinson Genetic Program (GP2). Unrelated to this manuscript, she received speaker honoraria from International Parkinson and Movement Disorders, Eisai and Orion Pharma and reports consultancies from Elsevier as Section Editor for Parkinsonism and Related Disorders. JT is supported by the German Research Foundation (DFG), Aligning Science Across Parkinson’s (ASAP) Global Parkinson’s Genetics Program (GP2), and is a consultant for Acurex. BT acknowledges support from the Global Parkinson’s Genetics Program (GP2). EMV is supported by the Aligning Science Across Parkinson’s (ASAP) Global Parkinson’s Genetics Program (GP2). Unrelated to this manuscript, she received research grants from Telethon Foundation Italy (GGP20070), the Italian Ministry of Health (Ricerca Finalizzata RF-2019-12369368 and ERA-NET Neuron NDCil project EUR002), the Italian Ministry of University and Research (GENERA, MNESYS) and the Silverstein Foundation. KL received grants from the Dystonia Medical Research Foundation and German Research Foundation (DFG), unrelated to this manuscript. CB is an employee of the Coalition for Aligning Science (CAS). AS is the lead investigator for a grant from the Michael J Fox Foundation for Parkinson’s Research and has a contract for work on the Global Parkinson’s Genetics Program (GP2). Unrelated to this manuscript, he is named as an inventor on patents for a diagnostic for stroke and for molecular testing for C9orf72 repeats; he is on the scientific advisory board of the Lewy Body Disease Association and for Cajal Neuroscience (both unpaid positions); and he received an honorarium for speaking at the World Laureates Association. HRM reports grant support from Parkinson’s UK, Cure Parkinson’s Trust, PSP Association, Medical Research Council, and the Michael J Fox Foundation. Unrelated to this manuscript, he is a co-applicant on a patent application related to C9orf72 - Method for diagnosing a neurodegenerative disease (PCT/GB2012/052140) and received honoraria from the Movement Disorder Society. CK received grants from the Michael J. Fox Foundation for Parkinson’s Research and the Aligning Science Across Parkinson’s Initiative. Unrelated to this manuscript, she received grants from the German Research Foundation and speakers’ honoraria from Bial. She has royalties at Oxford University Press and Springer Nature and serves as a medical advisor to Centogene and Biogen.
Figures


References
-
- Leonard HL, Global Parkinson’s Genetics Program (GP2). Novel Parkinson’s disease genetic risk factors within and across European populations. medRxiv. 2025; published online March 17. DOI: 10.1101/2025.03.14.24319455. - DOI
-
- Lange LM, Gonzalez-Latapi P, Rajalingam R, et al. Nomenclature of Genetic Movement Disorders: Recommendations of the International Parkinson and Movement Disorder Society Task Force - an update. Mov Disord 2022; 37: 905–35. - PubMed
-
- Lim S-Y, Tan AH, Ahmad-Annuar A, et al. Uncovering the genetic basis of Parkinson’s disease globally: from discoveries to the clinic. Lancet Neurol 2024; 23: 1267–80. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
