Characterization of inflammatory protein expression patterns and their association with viral DNA load in hepatitis B virus infection via Olink proteomics analysis
- PMID: 40672613
- PMCID: PMC12261205
- DOI: 10.62347/JTXH6594
Characterization of inflammatory protein expression patterns and their association with viral DNA load in hepatitis B virus infection via Olink proteomics analysis
Abstract
Objectives: To investigate the differential expression of inflammatory proteins in the sera of patients with chronic hepatitis B (CHB) using Olink Targeted Proteomics and to explore their correlations with viral deoxyribonucleic acid (DNA) load.
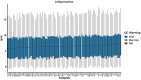
Methods: This retrospective study included 66 CHB patients and 22 healthy controls, with medical records collected between January and December 2023 at Nanning Customs, China. Viral DNA loads were quantified using real-time polymerase chain reaction (PCR), and 92 inflammatory proteins were profiled using Olink Targeted Proteomics.
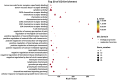
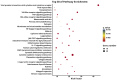
Results: A total of 38 proteins were differentially expressed between CHB patients and healthy controls, of which 7 were upregulated and 31 were downregulated. Correlation analysis revealed that viral DNA load was positively associated with the expression of osteoprotegerin (OPG) (P = 0.021) and chemokine (C-X-C motif) ligand 9 (CXCL9) (P = 0.002), and negatively associated with interleukin-10 (IL-10) (P = 0.007), cluster of differentiation 40 (CD40) (P = 0.004), and caspase-8 (CASP8) (P = 0.020). Functional enrichment analysis indicated that these proteins were mainly enriched in neutrophil chemotaxis, granulocyte migration, chemokine signaling pathways, cytokine activity, chemokine activity receptors, and tumor necrosis factor (TNF) receptors.
Conclusions: Specific inflammatory proteins, including OPG, CXCL9, IL-10, CD40, CASP8, are associated with viral DNA load in CHB patients. These findings enhance the proteomic understanding of HBV pathogenesis and may offer potential therapeutic targets and biomarkers.
Keywords: Hepatitis B virus; bioinformatics analysis; inflammatory factors; proteomics.
AJTR Copyright © 2025.
Conflict of interest statement
None.
Figures

Similar articles
-
Single-cell proteomics analysis of human oocytes during GV-to-MI transition.Hum Reprod. 2025 Jul 1;40(7):1332-1343. doi: 10.1093/humrep/deaf086. Hum Reprod. 2025. PMID: 40359387
-
C-X-C chemokine receptor type 5+CD8+ T cells as immune regulators in hepatitis Be antigen-positive chronic hepatitis B under interferon-alpha treatment.World J Gastroenterol. 2025 Jan 21;31(3):99833. doi: 10.3748/wjg.v31.i3.99833. World J Gastroenterol. 2025. PMID: 39839901 Free PMC article. Clinical Trial.
-
Adefovir dipivoxil and pegylated interferon alfa-2a for the treatment of chronic hepatitis B: a systematic review and economic evaluation.Health Technol Assess. 2006 Aug;10(28):iii-iv, xi-xiv, 1-183. doi: 10.3310/hta10280. Health Technol Assess. 2006. PMID: 16904047
-
Correlation between virological response and portal vein thrombosis in patients with chronic hepatitis B.Sci Rep. 2025 Jul 1;15(1):20856. doi: 10.1038/s41598-025-04316-6. Sci Rep. 2025. PMID: 40595918 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
References
-
- Robinson A, Wong R, Gish RG. Chronic hepatitis B virus and hepatitis D virus: new developments. Clin Liver Dis. 2023;27:17–25. - PubMed
-
- Xiao L, Tang K, Fu T, Yuan X, Seery S, Zhang W, Ji Z, He Z, Yang Y, Zhang W, Jia W, Liang C, Tang H, Wang F, Ye Y, Chen L, Shao Z. Cytokine profiles and virological markers highlight distinctive immune statuses, and effectivenesses and limitations of NAs across different courses of chronic HBV infection. Cytokine. 2024;173:156442. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
