A strain defined as a novel species in the Acinetobacter genus co-harboring chromosomal associated tet(X3) and plasmid associated bla NDM-1 from a beef cattle farm in Hebei, China
- PMID: 40673007
- PMCID: PMC12263697
- DOI: 10.3389/fcimb.2025.1594982
A strain defined as a novel species in the Acinetobacter genus co-harboring chromosomal associated tet(X3) and plasmid associated bla NDM-1 from a beef cattle farm in Hebei, China
Abstract
Introduction: The co-existence phenomenon of antibiotic resistance genes (ARGs), particularly of last-resort antibiotics in multi-drug resistant (MDR) bacteria, is of particular concern in the least studied bacterial species.
Methods: In 2023, strain M2 was isolated from the sludge sample at a commercial bovine farm in Hebei province, China, using a MacConkey plate containing meropenem. PCR amplification and Sanger sequencing verified it co-carrying bla NDM and tet(X) genes. It was classified within the Acinetobacter genus by MALDI-TOF-MS and 16S rDNA analyses. Whole-genome sequencing (WGS) was performed on the Oxford Nanopore platform, with species-level identification via ANI and dDDH. Antimicrobial susceptibility testing was performed against 20 antibiotics. Conjugation assays employed the filter-mating method using E. coli J53 and Salmonella LGJ2 as recipients.
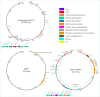
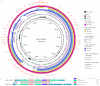
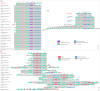
Results: This strain was confirmed as a novel species of Acinetobacter genus, showing resistance to meropenem, ampicillin, ceftazidime, cefepime, gentamicin, kanamycin, fosfomycin, imipenem, ertapenem, and tetracycline. Despite carrying tet(X3), it remained susceptible to tigecycline, omadacycline, and doxycycline. The genome carried 11 ARG types, multiple metal resistance genes (MRGs), and virulence factor (VF) genes. The bla NDM-1 was located in a skeleton, ISAba125-bla NDM-1-ble MBL-trpF, which was carried by an ISAba14-mediated rolling-circle-like structure in pM2-2-NDM-1 (rep_cluster_481). Integrative and conjugative element (ICE) and multiple pdif modules (driven by the XerCD site-specific recombination (XerCD SSR) system), which were associated with the mobilization of resistance determinants, were identified in this plasmid. Chromosomal tet(X3) was mediated by ISVsa3, forming a skeleton, ISVsa3-XerD-tet (X3)-res-ISVsa3.
Discussion: The co-occurrence of bla NDM and tet(X) in a novel species of the Acinetobacter genus hints that substantial undiscovered bacteria co-carrying high-risk ARGs are concealing in the agroecological system, which should cause particular concern.
Keywords: Acinetobacter; antibiotic resistance genes (ARGs); blaNDM; multi-drug resistance; pdif module; tet(X).
Copyright © 2025 Wang, Wei, Shoaib, Qiu, Zhang, Dai, Lin, Wang and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Diverse Acinetobacter in retail meat: a hidden vector of novel species and antimicrobial resistance genes, including plasmid-borne blaOXA-58, mcr-4.3 and tet(X3).Int J Food Microbiol. 2025 Oct 2;441:111313. doi: 10.1016/j.ijfoodmicro.2025.111313. Epub 2025 Jun 10. Int J Food Microbiol. 2025. PMID: 40513431
-
Whole genome analysis reveals the distribution and diversity of plasmid reservoirs of NDM and MCR in commercial chicken farms in China.Microbiol Spectr. 2025 Jul;13(7):e0290024. doi: 10.1128/spectrum.02900-24. Epub 2025 Jun 9. Microbiol Spectr. 2025. PMID: 40488461 Free PMC article.
-
Genomic characterization of plasmids harboring blaNDM-1, blaNDM-5, and blaNDM-7 carbapenemase alleles in clinical Klebsiella pneumoniae in Pakistan.Microbiol Spectr. 2025 Jul;13(7):e0235924. doi: 10.1128/spectrum.02359-24. Epub 2025 May 22. Microbiol Spectr. 2025. PMID: 40401976 Free PMC article.
-
Global Epidemiology and Antimicrobial Resistance of Metallo-β-Lactamase (MBL)-Producing Acinetobacter Clinical Isolates: A Systematic Review.Pathogens. 2025 Jun 3;14(6):557. doi: 10.3390/pathogens14060557. Pathogens. 2025. PMID: 40559565 Free PMC article. Review.
-
Prevalence of antibiotic resistance genes in the oral cavity and mobile genetic elements that disseminate antimicrobial resistance: A systematic review.Mol Oral Microbiol. 2022 Aug;37(4):133-153. doi: 10.1111/omi.12375. Epub 2022 Jun 17. Mol Oral Microbiol. 2022. PMID: 35674142
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

