Identification of a novel hypovirulence-inducing ourmia-like mycovirus from Fusarium solani causing ginseng (Panax ginseng) root rot
- PMID: 40673145
- PMCID: PMC12263584
- DOI: 10.3389/fmicb.2025.1609431
Identification of a novel hypovirulence-inducing ourmia-like mycovirus from Fusarium solani causing ginseng (Panax ginseng) root rot
Abstract
Introduction: Fusarium solani is a widespread plant pathogen known to damage numerous crops, including causing severe root rot in Panax ginseng. In this study, we identified a novel ourmia-like mycovirus in F. solani, named "Fusarium solani ourmia-like virus 1" (FsoOLV1). We demonstrated that FsoOLV1 confers hypovirulence in its host F. solani and three other ginseng root rot pathogens, F. oxysporum, F. proliferatum, and F. verticillioides. Additionally, we verified its horizontal and vertical transmission capabilities.
Methods: FsoOLV1 was discovered in F. solani strain SJH 2-4 using high-throughput sequencing. The genome sequence of FsoOLV1 was obtained through RT-PCR and RACE. Virus elimination was conducted to assess the effect of FsoOLV1 on fungal virulence. Protoplast transfection experiments were performed to evaluate the impact of the virus on other ginseng root rot pathogens. Horizontal and vertical transmission studies were also carried out to examine the spread of the virus.
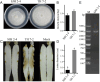
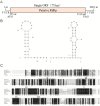
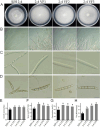
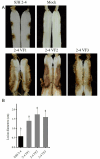
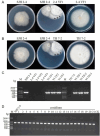
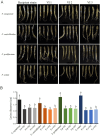
Results: The genome of FsoOLV1 is 2,801 nucleotides (nt) in length with a GC content of 47.05%. It encodes a 750 amino acid RNA-dependent RNA polymerase (RdRp), with a molecular weight of approximately 84.84 kDa. Phylogenetic analysis indicated that FsoOLV1 clusters within a clade containing the Magoulivirus genus in the Botourmiaviridae family. Curing FsoOLV1 from the fungal host strain revealed that the mycovirus plays a role in reducing the virulence of the F. solani strain SJH 2-4. Furthermore, protoplast transfection revealed that FsoOLV1 can significantly reduce the virulence of other ginseng root rot pathogens, including F. oxysporum, F. proliferatum, and F. verticillioides. Additionally, FsoOLV1 is capable of horizontal transmission between F. solani strains and vertical transmission to the next generation via conidia.
Discussion: This study presents the first hypovirulent ourmia-like virus, FsoOLV1, which reduces the virulence of F. solani and other ginseng root rot pathogens. Our findings suggest that FsoOLV1 could serve as a promising biological agent for controlling ginseng root rot. This research not only expands the diversity of known hypovirulent mycoviruses but also provides a potential candidate for controlling Fusarium diseases in ginseng cultivation.
Keywords: Fusarium solani; ginseng root rot; hypovirulence; mycovirus; ourmia-like virus.
Copyright © 2025 Ma, Ni, Liu, Cai, Jiang, Lu, Yang, Zhang and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Changes in the diversity of ginseng endophyte flora driven by Fusarium solani.Front Microbiol. 2025 Apr 28;16:1554706. doi: 10.3389/fmicb.2025.1554706. eCollection 2025. Front Microbiol. 2025. PMID: 40356640 Free PMC article.
-
Fungal Pathogens Associated with Crown and Root Rot in Wheat-Growing Areas of Northern Kyrgyzstan.J Fungi (Basel). 2023 Jan 16;9(1):124. doi: 10.3390/jof9010124. J Fungi (Basel). 2023. PMID: 36675945 Free PMC article.
-
Identification, Pathogenicity, and Genetic Diversity of Fusarium spp. Associated with Maize Sheath Rot in Heilongjiang Province, China.Int J Mol Sci. 2022 Sep 16;23(18):10821. doi: 10.3390/ijms231810821. Int J Mol Sci. 2022. PMID: 36142733 Free PMC article.
-
123I-MIBG scintigraphy and 18F-FDG-PET imaging for diagnosing neuroblastoma.Cochrane Database Syst Rev. 2015 Sep 29;2015(9):CD009263. doi: 10.1002/14651858.CD009263.pub2. Cochrane Database Syst Rev. 2015. PMID: 26417712 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

