Aldehyde Oxidase 1 Deficiency Enhances Aerobic Exercise Performance by Promoting Skeletal Muscle Adaptation and Improving Mitochondrial Function
- PMID: 40673877
- PMCID: PMC12269804
- DOI: 10.1096/fj.202500240R
Aldehyde Oxidase 1 Deficiency Enhances Aerobic Exercise Performance by Promoting Skeletal Muscle Adaptation and Improving Mitochondrial Function
Abstract
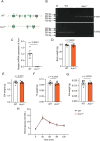
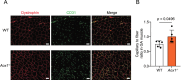
Aerobic exercise has significant health benefits, including preventing chronic diseases like sarcopenia. It strongly depends on muscle fiber types, with higher oxidative fiber ratios enhancing endurance. However, the molecular mechanisms underlying aerobic exercise capacity remain incompletely understood. In this study, we identified 395 genes associated with muscle fiber types, among which 39 were linked to metabolic pathways. Notably, we focused on aldehyde oxidase 1 (AOX1), a molybdenum flavin enzyme, due to its unique non-mitochondrial localization, suggesting a potential causal role in regulating muscle metabolism. We further revealed a significant downregulation of Aox1 mRNA expression in the skeletal muscle of mice after two weeks of exercise training, indicating its involvement in exercise adaptation. To further explore this link, we generated Aox1 knockout (KO) mice and subjected them to endurance capacity tests. Aox1 KO mice exhibited significantly enhanced exercise endurance compared to wild-type (WT) controls, accompanied by a shift toward a more oxidative muscle phenotype, as indicated by an increased proportion of oxidative fibers. Mechanistically, Aox1 KO mice exhibit increased expression of PGC-1α, enhanced mitochondrial function, and increased capillary density in skeletal muscle, facilitating improved oxygen delivery and utilization during exercise. Additionally, in vitro experiments using C2C12 myotubes revealed that Aox1 knockdown alleviated starvation- and TNF-α-induced muscle atrophy, which partially mimics sarcopenia, highlighting its protective role against aging- and stress-induced muscle damage. These findings identify AOX1 as a negative regulator of aerobic exercise capacity and stress resilience, advancing our understanding of skeletal muscle adaptation and highlighting AOX1 as a potential target for improving exercise performance and mitigating sarcopenia.
Keywords: AOX1; aerobic exercise; capillary density; mitochondria; sarcopenia; skeletal muscle.
© 2025 The Author(s). The FASEB Journal published by Wiley Periodicals LLC on behalf of Federation of American Societies for Experimental Biology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
MeSH terms
Substances
Grants and funding
- 2023YFC3603300/MOST | NSFC | Key Programme
- 2023YFF1001000/MOST | NSFC | Key Programme
- 82471591/MOST | National Natural Science Foundation of China (NSFC)
- 82460283/MOST | National Natural Science Foundation of China (NSFC)
- 20242BAB26156/Natural Science Foundation of Jiangxi Province (Jiangxi Natural Science Foundation)
LinkOut - more resources
Full Text Sources
Research Materials

