Mass Spectrometry Imaging
- PMID: 40674148
- PMCID: PMC12311901
- DOI: 10.1021/acs.analchem.4c05249
Mass Spectrometry Imaging
Abstract
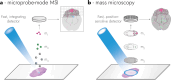
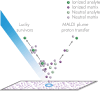
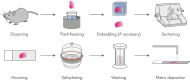
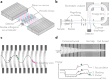
Mass spectrometry imaging (MSI) maps the spatial distributions of chemicals on chemically complex surfaces. MSI offers unrivaled sensitivity and information density with each pixel comprising a mass spectrum. Over the past three decades, numerous technological developments have enabled MSI to evolve into a mainstream technique for untargeted molecular and elemental imaging with wide-spread applications ranging from material analysis to life sciences and clinical diagnostics. Here, we review the field of MSI with a focus on key technological advancements. We examine different image acquisition modes and the most popular ionization methods in MSI, including matrix-assisted laser desorption/ionization (MALDI), laser ablation inductively coupled plasma (LA-ICP), laser ablation electrospray ionization (LAESI), secondary ion mass spectrometry (SIMS), and desorption electrospray ionization (DESI). For each method, we discuss figures of merit, such as spatial resolving power and sensitivity, the ionization mechanism, sample preparation, advantages, and disadvantages, including ways to overcome them wherever applicable. We subsequently discuss more aspects of MSI instrumentation, such as commonly used mass analyzers, tandem mass spectrometry, ion mobility, and advancements in imaging throughput. Based on these technological developments, targeted MSI strategies are explained, including imaging mass cytometry (IMC), multiplexed ion beam imaging (MIBI), and stable isotope labeling (SIL), as well as approaches for multimodal imaging. Last, we present selected application examples of MSI in cancer research, single cell analysis, and drug distribution studies. We target this review to provide researchers with an interest in recent developments in MSI with a concise technological understanding of the different main approaches to MSI.
Figures







References
-
- Maric D., Jahanipour J., Li X. R., Singh A., Mobiny A., van Nguyen H., Sedlock A., Grama K., Roysam B.. Whole-Brain Tissue Mapping Toolkit Using Large-Scale Highly Multiplexed Immunofluorescence Imaging and Deep Neural Networks. Nat Commun. 2021;12(1):1–12. doi: 10.1038/s41467-021-21735-x. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

