Membrane-wide screening identifies potential tissue-specific determinants of SARS-CoV-2 tropism
- PMID: 40674406
- PMCID: PMC12286382
- DOI: 10.1371/journal.ppat.1013157
Membrane-wide screening identifies potential tissue-specific determinants of SARS-CoV-2 tropism
Abstract
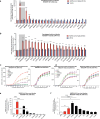
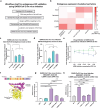
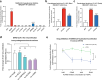
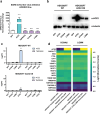
While SARS-CoV-2 primarily infects the respiratory tract, clinical evidence indicates that cells from diverse cell types and organs are also susceptible to infection. Using the CRISPR activation (CRISPRa) approach, we systematically targeted human membrane proteins in cells with and without overexpression of ACE2, thus identifying unrecognized host factors that may facilitate viral entry. Validation experiments with replication-competent SARS-CoV-2 confirmed the role of newly identified host factors, particularly the endo-lysosomal protease legumain (LGMN) and the potassium channel KCNA6, upon exogenous overexpression. In orthogonal experiments, we show that disruption of endogenous LGMN or KCNA6 decreases viral infection and that inhibitors of candidate factors can reduce viral entry. Additionally, using clinical data, we find possible associations between expression of either LGMN or KCNA6 and SARS-CoV-2 infection in human tissues. Our results identify potentially druggable host factors involved in SARS-CoV-2 entry, and demonstrate the utility of focused, membrane-wide CRISPRa screens in uncovering tissue-specific entry factors of emerging pathogens.
Copyright: © 2025 Dinesh et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Mechanisms of SARS-CoV-2 Transmission and Pathogenesis - PubMed. [cited 9 May 2025]. Available from: https://pubmed.ncbi.nlm.nih.gov/33132005/
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

