Interferon gamma rebalances immunopathological signatures in chronic granulomatous disease through metabolic rewiring
- PMID: 40674716
- PMCID: PMC12556223
- DOI: 10.1182/bloodadvances.2025016213
Interferon gamma rebalances immunopathological signatures in chronic granulomatous disease through metabolic rewiring
Abstract
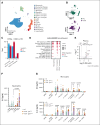
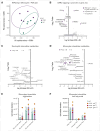
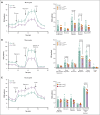
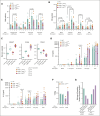
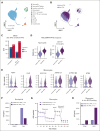
Chronic granulomatous disease (CGD) is a primary immunodeficiency characterized by recurrent life-threatening infections and hyperinflammatory complications. It is caused by mutations in the reduced nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex and the consequent loss of reactive oxygen species (ROS) production. Recombinant human interferon gamma (rIFN-γ) prophylaxis reduces the risk of severe infections, but the mechanisms behind its efficacy in CGD are still an open question, as it does not restore NADPH oxidase-dependent ROS production. Here, we demonstrate that the innate immune cells of patients with CGD are transcriptionally and functionally reprogrammed to a hyperactive inflammatory status, displaying an impaired in vitro induction of trained immunity. CGD monocytes have reduced intracellular amino acid concentrations and profound functional metabolic defects, both at the level of glycolysis and mitochondrial respiration. Ex vivo and in vivo treatments with IFN-γ restored these metabolic defects and reduced excessive interleukin 1β (IL-1β) and IL-6 production in response to fungal stimuli in CGD monocytes. These data suggest that prophylactic rIFN-γ modulates the metabolic status of innate immune cells in CGD. These data shed light on the effects of NADPH oxidase-derived ROS deficiency to the metabolic programs of immune cells and pose the basis for targeting this immunometabolic axis, potentially beyond CGD, with IFN-γ immunotherapy.
© 2025 American Society of Hematology. Published by Elsevier Inc. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: M.S. was the inventor of the TruCulture technology and is the CEO of HOT Screen GmbH, the company that manufactures TruCulture. The remaining authors declare no competing financial interests.
Figures




References
-
- Schuett J, Schuett H, Oberoi R, et al. NADPH oxidase NOX2 mediates TLR2/ 6-dependent release of GM-CSF from endothelial cells. The FASEB J. 2017;31(6):2612–2624. - PubMed
-
- Seger RA. Chronic granulomatous disease 2018: advances in pathophysiology and clinical management. LymphoSign J. 2019;6(1):1–16.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

