Genomics of chronic dry cough unravels neurological pathways
- PMID: 40675770
- PMCID: PMC12461901
- DOI: 10.1183/13993003.02341-2024
Genomics of chronic dry cough unravels neurological pathways
Abstract
Background: Chronic dry cough is a symptom of common lung conditions, can occur as a side-effect of angiotensin-converting enzyme inhibitors (ACEis), or may be unexplained. Despite the substantial health burden presented by chronic dry cough, its biological mechanisms remain unclear. We hypothesised shared genetic architecture between chronic dry cough and ACEi-induced cough and aimed to identify causal genes underlying both phenotypes.
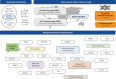
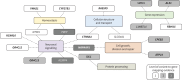
Methods: We performed multi-ancestry genome-wide association studies (GWAS) of chronic dry cough and ACEi-induced cough, and a multi-trait GWAS of both phenotypes, utilising data from five cohort studies. Chronic dry cough was defined by questionnaire responses, and ACEi-induced cough by treatment switches or clinical diagnosis in electronic health records. We mapped putative causal genes and performed phenome-wide association studies (PheWAS) of associated variants, and polygenic scores for ACEi-induced cough, to identify pleiotropic effects.
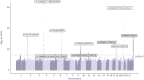
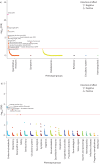
Results: We found seven novel genetic association signals reaching p<5×10-8 in the multi-trait or single-trait analyses of chronic dry cough and ACEi-induced cough. The novel variants mapped to 10 novel genes, and we mapped an additional three novel genes to known risk variants, many of which implicate neurological functions (CTNNA1, KCNA10, MAPKAP1, OR4C12, OR4C13, SIL1). The polygenic-score-based PheWAS highlighted associations with an elevated risk of several clinical conditions including asthma, diabetes and multi-site chronic pain.
Conclusion: Our findings provide support for neuronal dysfunction underlying cough hypersensitivity in chronic dry cough and ACEi-induced cough, and identify diseases and traits associated with genetic predisposition to cough that could inform drug target discovery.
Copyright ©The authors 2025.
Conflict of interest statement
Conflict of interest: C. John reports support for the present manuscript from Orion Pharma. R. Packer reports support for the present manuscript from Orion Pharma. C. Erikstrup reports grants from Novo Nordisk and Abbott Diagnostics. W. Hennah is an employee of Orion Pharma. M. Marttila is an employee of Orion Pharma. L.V. Wain reports support for the current manuscript from Orion Pharma, grants from GlaxoSmithKline, and consultation fees from Galapagos. M.D. Tobin reports support for the current manuscript from Orion Pharma, and research collaborations with GlaxoSmithKline. The remaining authors have no potential conflicts of interest to disclose.
Figures

Comment in
-
Dissecting the genetics of chronic cough.Eur Respir J. 2025 Sep 25;66(3):2501267. doi: 10.1183/13993003.01267-2025. Print 2025 Sep. Eur Respir J. 2025. PMID: 40998561 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
