Global dissemination of npmA mediated pan-aminoglycoside resistance via a mobile genetic element in Gram-positive bacteria
- PMID: 40675954
- PMCID: PMC12271512
- DOI: 10.1038/s41467-025-61152-y
Global dissemination of npmA mediated pan-aminoglycoside resistance via a mobile genetic element in Gram-positive bacteria
Abstract
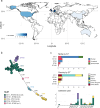
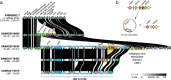
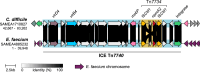
The npmA gene, encoding a 16S rRNA methyltransferase, confers resistance to all clinically available aminoglycosides, posing a significant threat to effective antibiotic therapy. We analyze 1,932,812 bacterial genomes to investigate the distribution and mobilization of npmA variants. npmA is not found in Gram-negative bacteria, where it was originally described, but is identified among Gram-positive bacteria, predominantly as the npmA2 variant in the globally distributed Clostridioides difficile ST11 lineage. We also detect npmA2 in two vancomycin-resistant Enterococcus faecium isolates from a Dutch hospital. Upon sequencing and phenotypic analysis, we determine that E. faecium isolates are pan-resistant to aminoglycosides. Genomic characterization links npmA2 to a composite transposon, Tn7734, which is integrated within a previously uncharacterized Integrative and Conjugative Element (ICE) Tn7740, present in both npmA2-carrying C. difficile and E. faecium clinical isolates. Tn7740-like, but not npmA2, appears across diverse taxa, including human microbiome members. Here, we show that Tn7740 likely facilitates cross-species npmA2 mobilization between these Gram-positive bacteria and emphasize the risk of mobile genetic elements transferring pan-aminoglycoside resistance between clinically important bacterial pathogens.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
-
- Zhanel, G. G. et al. Comparison of the next-generation aminoglycoside plazomicin to gentamicin, tobramycin and amikacin. Expert Rev. Anti Infect. Ther.10, 459–473 (2012). - PubMed
-
- Gür, D. et al. Comparative in vitro activity of plazomicin and older aminoglycosides against Enterobacterales isolates; prevalence of aminoglycoside modifying enzymes and 16S rRNA methyltransferases. Diagn. Microbiol. Infect. Dis.97, 115092 (2020). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

