Near-infrared light-driven metabolic reprogramming of synoviocytes for the treatment of rheumatoid arthritis
- PMID: 40676021
- PMCID: PMC12271504
- DOI: 10.1038/s41467-025-61923-7
Near-infrared light-driven metabolic reprogramming of synoviocytes for the treatment of rheumatoid arthritis
Abstract
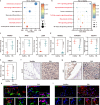
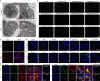
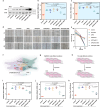
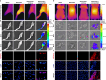
Rheumatoid arthritis is a common autoimmune disease characterized by chronic synovial inflammation and joint destruction, primarily driven by an imbalanced cellular metabolism and inflammatory microenvironment. While gene therapy offers a promising therapeutic approach, its effectiveness is limited by the challenges of non-specific gene expression in healthy tissues. Here, we develop a gene delivery system (namely APPC), in which near-infrared (NIR)-responsive gold nanorods are coated with chondroitin sulfate-modified polyethyleneimine to facilitate the heat-responsive targeted delivery of heme oxygenase 1 (HO-1) gene. The APPC shows favorable transfection efficiency due to its targeting ability and significantly facilitates HO-1 expression under NIR irradiation. The combination of APPC/pHO-1 and NIR can effectively reprogram the cellular metabolism and repolarize the macrophages and fibroblast-like synoviocytes, thereby inhibiting inflammation by suppressing glycolysis. Meanwhile, APPC can specifically enhance the HO-1 expression in inflamed tissues through NIR-mediated the activation of heat shock protein 70 promoter, ensuring the precise gene expression via photothermal conversion. In a collagen-induced arthritis model, APPC/pHO-1 under NIR irradiation exhibits potent therapeutic efficacy, restoring the articular microenvironmental homeostasis and mitigating the symptoms of rheumatoid arthritis. These findings highlight the potential of APPC/pHO-1 nanoparticles in the gene therapy of rheumatoid arthritis and other inflammatory diseases.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

