Disrupted Lipid Metabolism Aggravates Ischemic Brain Injury: Targeting FDFT1 for Stroke Therapy
- PMID: 40676368
- PMCID: PMC12511267
- DOI: 10.1007/s12035-025-05217-5
Disrupted Lipid Metabolism Aggravates Ischemic Brain Injury: Targeting FDFT1 for Stroke Therapy
Abstract
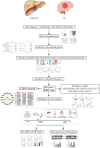
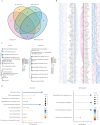
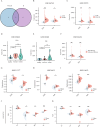
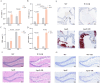
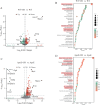
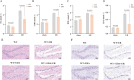
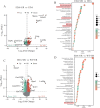
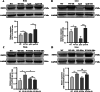
Lipid metabolism disorder has been established as a contributing factor to the exacerbation of ischemic stroke (IS) damage. Conditions such as metabolic dysfunction-associated steatotic liver disease (MASLD) and atherosclerosis are known to elevate the risk of IS. Therefore, elucidating the association between potential risk factors of IS and the pathogenesis of IS from the perspective of lipid regulation may provide new insight for the prevention and treatment. In our study, we obtained Gene Expression Omnibus Series (GSE) from NCBI (National Center for Biotechnology Information) GEO (Gene Expression Omnibus). Through the analysis of the datasets in MASLD and IS patients, we found that abnormal lipid metabolism is a potential pathway for stroke induced by MASLD as a risk factor. Furthermore, we established a middle cerebral artery occlusion-reperfusion (MCAO/R) model in mice, measured atherosclerotic lesions in ApoE-deficient mice, and performed RNA-seq analysis to identify differentially expressed genes (DEGs) following IS. Our findings indicate that the DEGs are associated with lipid metabolism signaling pathways and inflammatory response pathways. The ApoE mice exhibited more severe IS injury. edaravone, a free radical scavenger clinically used for acute ischemic stroke treatment, was employed here to investigate whether its neuroprotective pathways intersect with lipid metabolism regulation. We found that treatment with edaravone rectified metabolic disorders and mitigated IS damage. Furthermore, we observed that the expression of the hub gene Fdft1 was upregulated in both the brain and liver post-IS injury and significantly reduced following edaravone treatment. These findings suggest that lipid regulation is a promising avenue for IS therapy, and Fdft1 may emerge as a critical target for modulating lipid metabolism in the aftermath of IS.
Keywords: Atherosclerosis; Disrupted lipid metabolism; Fdft1; Ischemic stroke; MASLD.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics Approval: This study was approved by the Committee on Ethical Use of Animals of Chengdu University of Traditional Chinese Medicine (NO. 2023DL-022). Consent for Publication: Not applicable. Competing Interests: The authors declare no competing interests.
Figures


References
-
- Powers WJ (2020) Acute ischemic stroke. N Engl J Med 383:252–260 - PubMed
-
- Yang P, Zhang Y, Liu J (2025) Ischaemic stroke in 2024: progress on multiple fronts. Lancet. Neurol 24(1):7-8 - PubMed
-
- Catanese L, Tarsia J, Fisher M (2017) Acute ischemic stroke therapy overview. Circ Res 120:541–558 - PubMed
MeSH terms
Grants and funding
- 82305411/the National Natural Science Foundation of China
- 82430124/the National Natural Science Foundation of China
- 2023MD744131/Postdoctoral Fellowship Program of CPSF
- 2023NSFSC1822/the Department of Science and Technology of Sichuan Province
- 24NSFSC0097/the Department of Science and Technology of Sichuan Province
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

