Genome identification of NAC gene family and its gene expression patterns in responding to salt and drought stresses in Rhododendron delavayi
- PMID: 40676511
- PMCID: PMC12273022
- DOI: 10.1186/s12870-025-06965-1
Genome identification of NAC gene family and its gene expression patterns in responding to salt and drought stresses in Rhododendron delavayi
Abstract
Background: The NAC (NAM, ATAF1/2, and CUC2) transcription factors (TFs), which play a vital role in plant growth and development, stress response, and disease resistance, have been extensively analyzed in various plant species. However, there is limited knowledge regarding the NAC family in Rhododendron delavayi, an important ornamental flower.
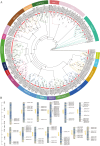
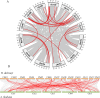
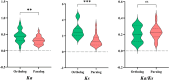
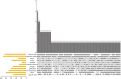
Result: In this study, a total of 102 RdNAC genes were identified from the R. delavayi genome. Phylogenetic analysis divided these genes into seven subfamilies, each characterized by similar conserved motifs. Chromosomal mapping revealed an uneven distribution of RdNACs across all 13 chromosomes, with gene family expansion driven primarily by dispersed duplication (110 gene pairs) and, to a lesser extent, tandem duplication (17 pairs). Intraspecific synteny analysis detected 26 pairs of duplicated RdNAC genes, while interspecific collinearity with Arabidopsis thaliana uncovered 83 orthologous pairs, indicating both lineage-specific diversification and conserved evolutionary relationships. Ka/Ks ratio calculations for both intra-RdNAC duplicates and RdNAC-AtNAC orthologs yielded values below 0.5, reflecting strong purifying selection. Conserved-motif and domain analyses identified ten distinct motifs and 35 structural domains, with the NAM and MIT CorA-like superfamily domains being the most prevalent. Promoter analysis of 2 kb upstream regions revealed a high abundance of abiotic stress-related cis-acting elements (e.g., ABRE, ARE, CGTCA-motif, LTR). Finally, qRT-PCR demonstrated that under drought (20% PEG) and salt (200 mM NaCl) treatments, multiple RdNACs, particularly RdNAC022 and RdNAC099, were significantly upregulated, underscoring their potential roles in stress response.
Conclusion: This study provides a comprehensive identification of the RdNAC TF family in R. delavayi, contributing to a better understanding of these transcription factors in this species. The findings also serve as a reference for analyzing stress responses, particularly concerning drought and salt stress in R. delavayi.
Keywords: NAC gene family; Rhododendron Delavayi; Abiotic stresses; Gene expression.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Rhododendron delavayi seedlings were collected from the Baili Rhododendron Nature Reserve in Guizhou and were identified by Professor Jing Ou. The methods involved in this study were carried out in compliance with local and national regulations. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
Comparative analysis of POD gene family and expression differentiation under NaCl, H2O2 and PEG stresses in sorghum.BMC Genomics. 2025 Jul 17;26(1):674. doi: 10.1186/s12864-025-11858-6. BMC Genomics. 2025. PMID: 40676507 Free PMC article.
-
Genome-wide identification of peanut ERFs and functional characterization of AhERF28 in response to salt and drought stresses.Plant Cell Rep. 2025 Jul 1;44(7):165. doi: 10.1007/s00299-025-03557-z. Plant Cell Rep. 2025. PMID: 40593149
-
[Identification of PLATZ gene family in Camellia sinensis and expression analysis of this gene family under high temperature and drought stresses].Sheng Wu Gong Cheng Xue Bao. 2025 Jul 25;41(7):2897-2912. doi: 10.13345/j.cjb.240969. Sheng Wu Gong Cheng Xue Bao. 2025. PMID: 40769568 Chinese.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
In silico analysis of the Seven IN Absentia (SINA) genes in bread wheat sheds light on their structure in plants.PLoS One. 2023 Dec 21;18(12):e0295021. doi: 10.1371/journal.pone.0295021. eCollection 2023. PLoS One. 2023. PMID: 38127955 Free PMC article.
References
-
- Olsen AN, Ernst HA, Leggio LL, Skriver K. NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 2005;10:79–87. - PubMed
-
- Kikuchi K, Ueguchi-Tanaka M, Yoshida KT, Nagato Y, Matsusoka M, Hirano HY. Molecular analysis of the NAC gene family in rice. Mol Gen Genet MGG. 2000;262:1047–51. - PubMed
-
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta BBA - Gene Regul Mech. 2012;1819:97–103. - PubMed
-
- Zhu B, Huo DA, Hong XX, Guo J, Peng T, Liu J, et al. The Salvia miltiorrhiza NAC transcription factor SmNAC1 enhances zinc content in transgenic Arabidopsis. Gene. 2019;688:54–61. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

