Explainable machine learning-driven models for predicting Parkinson's disease and its prognosis: obesity patterns associations and models development using NHANES 1999-2018 data
- PMID: 40676536
- PMCID: PMC12273281
- DOI: 10.1186/s12944-025-02664-w
Explainable machine learning-driven models for predicting Parkinson's disease and its prognosis: obesity patterns associations and models development using NHANES 1999-2018 data
Abstract
Background: Parkinson's disease (PD) is a prevalent neurodegenerative condition, the effect of obesity on PD remains controversial. We aimed to investigate the associations of obesity patterns on PD and all-cause mortality, while developing machine learning (ML)-driven predictive and prognostic models for PD.
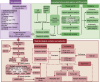
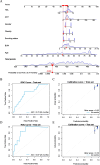
Methods: Fifty-one thousand, three hundred ninety-four adults from the National Health and Nutrition Examination Survey (NHANES) 1999-2018 were classified into four obesity patterns via body mass index (BMI) and waist circumference (WC). Associations of obesity patterns with PD risk and all-cause mortality were evaluated via multivariable logistic and Cox proportional hazards regression across three adjusted models. Subgroup, sensitivity, and restricted cubic spline (RCS) analyses examined stability, robustness, and nonlinearity. An integrative ML-driven architecture identified key features to develop predictive and prognostic nomograms, validated by the area under the receiver operating characteristic curves (AUCROCs) and calibration curves. Survival differences were analyzed using Kaplan-Meier curves. Shapley additive explanations (SHAP) enhanced model explanation.
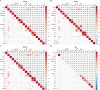
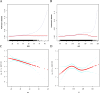
Results: Compound obesity significantly increased PD risk (Model 1: OR = 1.83, P < 0.001; Model 2: OR = 1.70, P = 0.002; Model 3: OR = 1.71, P = 0.006) yet correlated with reduced all-cause mortality in PD patients (Model 1: HR = 0.43, P = 0.003; Model 2: HR = 0.75, P = 0.428; Model 3: HR = 0.41, P = 0.033). Subgroup analysis revealed only HbA1c-modified association of compound obesity with PD (Pinteraction = 0.031). Sensitivity analyses confirmed robustness (pooled OR = 1.83, P < 0.001; pooled HR = 0.43, P = 0.003). RCS analyses revealed BMI-dependent PD risk escalation (Pnonlinearity = 0.008, BMI < 45.0 kg/m2), inverted U-shaped WC-PD link (Pnonlinearity < 0.001), and inverse dose-response BMI-mortality relationship (Pnonlinearity = 0.003), along with multiphasic WC-mortality association (PThreshold = 0.555 at 95 cm and PThreshold = 0.091 at 118 cm). LASSO + RF identified eight features, achieving moderate performance in PD prediction (SMOTE set: AUCROC = 0.75, Brier = 0.20) and prognosis (train set: AUCROC = 0.72, Brier = 0.22) nomograms, with similar results in the test set (AUCROC = 0.70, Brier = 0.01 for prediction, 0.87 and 0.18 for prognosis). No 24-month survival differences were observed across four obesity patterns (train set: Plog-rank = 0.73; test set: Plog-rank = 0.32).
Conclusions: This study preliminarily reveals that compound obesity significantly increases PD risk yet paradoxically associates with reduced all-cause mortality in PD patients. Validated predictive and prognostic nomograms for PD achieve relatively robust performances. Nonetheless, extensive longitudinal studies are required to validate these exploratory findings more comprehensively.
Keywords: Association; Multiple machine learning; Obesity pattern; Parkinson's disease; Prediction model; Shapley additive explanations.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The NCHS Ethics Review Board evaluated and granted approval for the NHANES survey ( https://www.cdc.gov/nchs/nhanes/irba98.htm ), and informed consent was obtained from all participants at the time of NHANES enrollment. Due to the nature of open access and the deidentification of data, this study was exempt from ethical approval by the Ethics Committee of Shaanxi Provincial People’s Hospital. Consent for publication: All authors have read, validate the accuracy of the data and approved the final manuscript. Competing interests: The authors declare no competing interests.
Figures






References
-
- Zamanian MY, Nazifi M, Khachatryan LG, et al. The neuroprotective effects of agmatine on Parkinson’s disease: focus on oxidative stress. Inflamm Mol Mech Inflamm. 2024. 10.1007/s10753-024-02139-7. - PubMed
-
- Jeong SM, Song YD, Seok CL, et al. Machine learning-based classification of Parkinson’s disease using acoustic features: insights from multilingual speech tasks. Comput Biol Med. 2024;182:109078. - PubMed
-
- Pringsheim T, Jette N, Frolkis A, et al. The prevalence of Parkinson’s disease: a systematic review and meta-analysis. Mov Disord. 2014;29(13):1583–90. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

