DNMT1 blocks SOX21-repressed CKS2 transcription to promote gastric cancer progression
- PMID: 40676553
- PMCID: PMC12273269
- DOI: 10.1186/s12885-025-14577-z
DNMT1 blocks SOX21-repressed CKS2 transcription to promote gastric cancer progression
Abstract
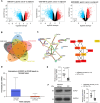
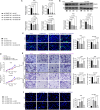
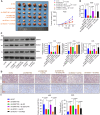
Background: The dysregulation of SOXs is related to tumor invasion, metastasis, proliferation, apoptosis, and epithelial-mesenchymal transition. This research sought to investigate the function and mechanisms of SOX21 in gastric cancer (GC).
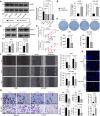
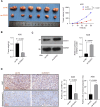
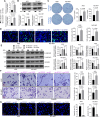
Methods: Multiple databases were included to determine the hub transcription factors in GC. In addition, RT-qPCR and Western blot were used to validate gene expression in tissues from GC patients. CCK-8, EdU, colony formation, wound healing, Transwell assays, and a xenograft tumor model were used to determine the function of SOX21 in GC. The targets of SOX21 were predicted and verified using ChIP, dual-luciferase reporter, and functional assays. SOX21 DNA methylation in GC cells was determined by qMSP. Rescue experiments were carried out in GC cells with DNMT1 silencing alone or in combination with SOX21 silencing.
Results: SOX21 was downregulated in GC tissues and cells. Ectopic expression of SOX21 inhibited cell growth, invasion, and migration, and induced apoptosis of GC cells. CKS2 was a target of SOX21, and overexpression of CKS2 promoted cell viability and mobility in GC cells overexpressing SOX21. The downregulation of SOX21 was related to the DNA hypermethylation catalyzed by DNMT1. The silencing of SOX21, by contrast, overturned the anti-tumor effects of sh-DNMT1 in vitro and in vivo.
Conclusion: Our data showed that DNMT1 overexpression upregulated CKS2 expression via hypermethylation of SOX21, thus promoting GC cell proliferation and growth, indicating that the DNMT1/SOX21/CKS2 axis could be a target for GC treatment.
Supplementary Information: The online version contains supplementary material available at 10.1186/s12885-025-14577-z.
Keywords: CKS2; DNMT1; Gastric cancer; Hypermethylation; SOX21.
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study was approved by the Ethics Committee of the Affiliated Chuzhou Hospital of Anhui Medical University. All patients offered signed written informed consent for participation. All experiments adhered to the principles of the Declaration of Helsinki. The animal experiments involved in this study were approved by the Animal Ethics Committee of the Affiliated Chuzhou Hospital of Anhui Medical University. Competing interests: The authors declare no competing interests. Consent for publication: Not applicable.
Figures


Similar articles
-
Upregulation of the cycline kinase subunit CKS2 increases cell proliferation rate in gastric cancer.J Cancer Res Clin Oncol. 2009 Jun;135(6):761-9. doi: 10.1007/s00432-008-0510-3. Epub 2008 Nov 26. J Cancer Res Clin Oncol. 2009. PMID: 19034516 Free PMC article.
-
Clinicopathological and biological significance of CDC28 protein kinase regulatory subunit 2 overexpression in human gastric cancer.Int J Oncol. 2011 Aug;39(2):361-72. doi: 10.3892/ijo.2011.1056. Epub 2011 May 25. Int J Oncol. 2011. PMID: 21617860
-
MicroRNA-148a is silenced by hypermethylation and interacts with DNA methyltransferase 1 in gastric cancer.Med Oncol. 2012 Dec;29(4):2701-9. doi: 10.1007/s12032-011-0134-3. Epub 2011 Dec 14. Med Oncol. 2012. PMID: 22167392
-
Long non-coding RNA SOX21-AS1: A potential tumor oncogene in human cancers.Pathol Res Pract. 2023 Sep;249:154774. doi: 10.1016/j.prp.2023.154774. Epub 2023 Aug 19. Pathol Res Pract. 2023. PMID: 37633003 Review.
-
Biological functions and therapeutic potential of CKS2 in human cancer.Front Oncol. 2024 Aug 12;14:1424569. doi: 10.3389/fonc.2024.1424569. eCollection 2024. Front Oncol. 2024. PMID: 39188686 Free PMC article. Review.
References
-
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;74(3):229–63. 10.3322/caac.21834. - PubMed
-
- Kim HD, Ryu MH, Kang YK. Adjuvant treatment for locally advanced gastric cancer: an Asian perspective. Gastric Cancer. 2024;27(3):439–50. 10.1007/s10120-024-01484-8. - PubMed
-
- Zeng Y, Jin RU. Molecular pathogenesis, targeted therapies, and future perspectives for gastric cancer. Semin Cancer Biol. 2022;86(Pt 3):566–82. 10.1016/j.semcancer.2021.12.004. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

