Ensemble learning for microbiome-based caries diagnosis: multi-group modeling and biological interpretation from salivary and plaque metagenomic data
- PMID: 40676575
- PMCID: PMC12272970
- DOI: 10.1186/s12903-025-06590-2
Ensemble learning for microbiome-based caries diagnosis: multi-group modeling and biological interpretation from salivary and plaque metagenomic data
Abstract
Background: Oral microbiota is a major etiological factor in the development of dental caries. Next-generation sequencing techniques have been widely used, generating vast amounts of data which is underexplored. The advancement of artificial intelligence (AI) technologies has made it possible to mine information from these large datasets. This study aimed to develop AI-driven diagnostic models and identify key microbial features for caries.
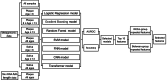
Methods: We collected raw metagenomic and full-length 16 S rRNA gene sequencing data from previous studies on saliva and plaque to construct a caries AI training dataset comprising nearly 600 samples. Samples were grouped based on age, sequencing and sampling method. Through systematic comparison of seven machine learning architectures, including Logistic Regression, Random Forest, Support Vector Machines, Gradient Boosting, Convolutional Neural Networks, Feedforward Neural Networks, and Transformer models, we developed subgroup-specific caries diagnostic models, with subsequent ensemble learning integration to enhance generalizability.
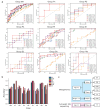
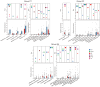
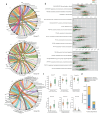
Results: The caries diagnostic model achieved a maximum AUC value of 1 (accuracy of 100%) for children under 6 years old in both saliva and plaque groups. The consistency of top features (species and metabolic pathways) contributing to the models was demonstrated through intra- and inter-group analyses. Key caries-associated species included Streptococcus salivarius, Streptococcus parasanguinis and Veillonella dispar. Veillonella parvula exhibits higher abundance in caries plaque samples, while being elevated in healthy saliva samples. Metabolic pathways like geranylgeranyl diphosphate and fructan biosynthesis were enriched in caries, whereas Bifidobacterium shunt and peptidoglycan biosynthesis were depleted.
Conclusion: The current work provided reliable diagnostic models for early childhood caries, and established a robust computational framework for AI-driven microbiome analysis. This study, by focusing on the characteristics of the oral microbiome, offers novel perspectives for data mining and validation of existing data through the application of AI modelling.
Keywords: Artificial intelligence; Early childhood caries; Metagenomics; Modelling; Salivary diagnostics.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This study involved a secondary analysis of publicly available, de-identified metagenomic and full-length 16S rRNA gene sequencing data. Therefore, ethical approval from our institutional review board was not required for this specific study. We confirm that all original studies from which the data were sourced obtained the necessary ethical approvals and informed consent from participants prior to data deposition in public repositories. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests. Clinical trial number: Not applicable.
Figures

References
-
- Bawaskar HS, Bawaskar PH. Oral diseases: a global public health challenge. Lancet (London England). 2020;395(10219):185–6. - PubMed
-
- Pitts NB, Zero DT, Marsh PD, Ekstrand K, Weintraub JA, Ramos-Gomez F, et al. Dental caries. Nat Reviews Disease Primers. 2017;3:17030. - PubMed
-
- Freire M, Nelson KE, Edlund A. The oral host-microbial interactome: an ecological chronometer of health? Trends Microbiol. 2021;29(6):551–61. - PubMed
-
- Simón-Soro A, Mira A. Solving the etiology of dental caries. Trends Microbiol. 2015;23(2):76–82. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

