Identification of prognosis and therapy related intratumoral microbiome and immune signatures in gastric cancer
- PMID: 40677704
- PMCID: PMC12267262
- DOI: 10.3389/fimmu.2025.1622959
Identification of prognosis and therapy related intratumoral microbiome and immune signatures in gastric cancer
Abstract
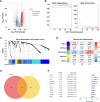
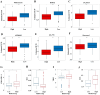
The impact of the intratumoral microbiome (ITM) on the treatment and prognosis of gastric cancer (GC) remains controversial. Our study analyzed the differential ITM in GC tissues and identified nine bacterial genera significantly associated with overall survival (OS), with seven as risk factors and two as protective factors. Three distinct clusters with varying survival outcomes were defined, demonstrating correlations with pathological stage and immune features. An immune-related gene-based RiskScore model incorporating genes such as Apolipoprotein D (APOD), Stanniocalcin 1 (STC1), Coagulation Factor II Thrombin Receptor (F2R), Angiotensinogen (AGT), Fatty Acid Binding Protein 4 (FABP4), Inhibin Subunit Beta A (INHBA), Caspase Recruitment Domain Family Member 11 (CARD11), and Dickkopf WNT Signaling Pathway Inhibitor 1 (DKK1) was established and validated in The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) datasets. When combined with clinical factors, this RiskScore model formed a Nomogram model achieving Areas Under the Curve (AUCs) of 0.72, 0.76, and 0.79 for 1, 3, and 5-year OS predictions, respectively. This model exhibited robust predictive accuracy over time and correlated with mutation frequency, drug sensitivity, and immunotherapy response. Furthermore, single-cell analysis revealed that tumor-associated fibroblasts may play a pivotal role in immune-microbial interactions. The results were confirmed using quantitative real-time polymerase chain reaction (qPCR) and immunohistochemistry (IHC). In conclusion, the prognostic model incorporating ITM and immune-related genes aids in risk stratification and provides valuable insights and targets for GC treatment.
Keywords: comprehensive analysis; drug sensitivity; gastric cancer; intratumoral microbiome; prognostic biomarkers; therapeutic responses.
Copyright © 2025 Jin, Ji, Bai, Su, Ke, Liu and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Predictive significance of MPT-driven necrosis-related genes signature in gastric cancer and their impact on the tumor microenvironment.Clin Transl Oncol. 2025 Jul;27(7):3015-3028. doi: 10.1007/s12094-024-03832-7. Epub 2024 Dec 17. Clin Transl Oncol. 2025. PMID: 39690336
-
Development and validation of an ARID1A-related immune genes risk model in evaluating prognosis and immune therapeutic efficacy for gastric cancer patients: a translational study.Front Immunol. 2025 Apr 28;16:1541491. doi: 10.3389/fimmu.2025.1541491. eCollection 2025. Front Immunol. 2025. PMID: 40406134 Free PMC article.
-
A novel copper-induced cell death-related lncRNA prognostic signature associated with immune infiltration and clinical value in gastric cancer.J Cancer Res Clin Oncol. 2023 Sep;149(12):10543-10559. doi: 10.1007/s00432-023-04916-7. Epub 2023 Jun 8. J Cancer Res Clin Oncol. 2023. PMID: 37291405 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

