TREM1 is essential for maintaining stemness of liver cancer stem-like cells in hepatocellular carcinoma
- PMID: 40677705
- PMCID: PMC12267286
- DOI: 10.3389/fimmu.2025.1618342
TREM1 is essential for maintaining stemness of liver cancer stem-like cells in hepatocellular carcinoma
Abstract
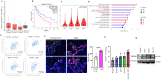
Introduction: Hepatocellular carcinoma (HCC) is the most common primary liver cancer and a leading cause of cancer-related mortality worldwide. While the Triggering Receptor Expressed on Myeloid Cells 1 (TREM1) is well-known for its role in amplifying inflammation within the tumor microenvironment (TME), its tumor-intrinsic role remains poorly defined. Liver cancer stem-like cells (LCSLCs), charecerized by expression of CD133 and EpCAM, are critical for HCC initiation, metastasis, recurrence, and therapy resistance.
Methods: We used flow cytometry to assess TREM1 expression in LCSLCs and employed CRISPR-Cas9 gene editing to knock out TREM1 in HCC cell lines. Functional assays, including proliferation, migration, apoptosis, clonogenicity, and spheroid formation, were performed. Cell line-derived xenograft (CDX) models were used to evaluate in vivo tumorigenicity. Transcriptomic profiling was conducted to explore downstream effects of TREM1 deletion. Additionally, a pharmacological inhibitor of TREM1 (VJDT) was used to validate the therapeutic potential of targeting TREM1 in vivo.
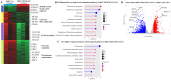
Results: TREM1 was highly expressed in CD133+EpCAM+ LCSLCs. Knockout of TREM1 significantly impaired proliferation and migration while promoting apoptosis in HCC cells. In LCSLCs, TREM1 silencing reduced clonogenic ability and spheroid formation, indicating loss of self-renewal and stemness. In CDX models, TREM1-deficient LCSLCs exhibited markedly reduced tumorigenicity. Transcriptomic analysis revealed distinct, context-dependent gene expression changes in nuclear and extracellular signaling pathways following TREM1 loss. Pharmacologic inhibition of TREM1 with VJDT recapitulated the tumor-suppressive effects observed in genetic models.
Discussion: Our findings establish TREM1 as a critical tumor-intrinsic regulator of LCSLC survival and tumorigenic potential, independent of its known immunomodulatory role in the TME. Targeting TREM1 may therefore represent a promising dual-action therapeutic strategy to disrupt both cancer stem-like cell function and the pro-inflammatory tumor milieu in HCC.
Keywords: TREM1; cancer stem cells; chemotherapy; hepatocellular carcinoma; oncogenesis.
Copyright © 2025 Sreekumar, Ajith, Mamouni, Horuzsko and Horuzsko.
Conflict of interest statement
AA and AH are inventors on a patent application (US patent no.US21/51072) submitted by Augusta University that covers compositions and methods for inhibiting TREM1. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

