ganon2: up-to-date and scalable metagenomics analysis
- PMID: 40677913
- PMCID: PMC12267982
- DOI: 10.1093/nargab/lqaf094
ganon2: up-to-date and scalable metagenomics analysis
Abstract
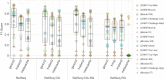
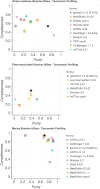
The fast growth of public genomic sequence repositories greatly contributes to the success of metagenomics. However, they are growing at a faster pace than the computational resources to use them. This challenges current methods, which struggle to take full advantage of massive and fast data generation. We propose a generational leap in performance and usability with ganon2, a sequence classification method that performs taxonomic binning and profiling for metagenomics analysis. It indexes large datasets with a small memory footprint, maintaining fast, sensitive, and precise classification results. Based on the full NCBI RefSeq and its subsets, ganon2 indices are on average 50% smaller than state-of-the-art methods. Using 16 simulated samples from various studies, including the CAMI 1+2 challenge, ganon2 achieved up to 0.15 higher median F1-score in taxonomic binning. In profiling, improvements in the F1-score median are up to 0.35, keeping a balanced L1-norm error in the abundance estimation. ganon2 is one of the fastest tools evaluated and enables the use of larger, more diverse, and up-to-date reference sets in daily microbiome analysis, improving the resolution of results. The code is open-source and available with documentation at https://github.com/pirovc/ganon.
© The Author(s) 2025. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Conflict of interest statement
None declared.
Figures




References
-
- GenBank and WGS statistics. (6 September 2023, date last accessed)https://www.ncbi.nlm.nih.gov/genbank/statistics/.
-
- DNA sequencing costs: data. (6 September 2023, date last accessed)https://www.genome.gov/about-genomics/fact-sheets/DNA-Sequencing-Costs-Data.
MeSH terms
LinkOut - more resources
Full Text Sources

