Species-level enumeration of low-concentration lactic acid bacteria in breast milk
- PMID: 40678022
- PMCID: PMC12270009
- DOI: 10.1016/j.fochms.2025.100269
Species-level enumeration of low-concentration lactic acid bacteria in breast milk
Abstract
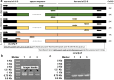
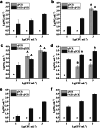
It is crucial to identify the distribution of lactic acid bacteria (LAB) in breast milk which is a source of live probiotics for the infant gut. Quantitative PCR (qPCR) is a powerful technique for selectively counting LAB at the species or strain level. However, the limit of quantification (LoQ) of qPCR is inadequate for human milk samples. To address this issue, nested primer pairs for Lactobacillus and Bifidobacterium species were designed to enable 15 cycles of preamplification Subsequent qPCR assays using the preamplification products (with 10-fold dilution) as templates reduced the LoQ for enumeration plasmids to one-thousandth of the original concentration. Importantly, preamplification did not enhance the detection capability for low biomass samples because the efficiency of DNA extraction was too low. To mitigate the variation in bacteria concentration, inert bacteria were added to the samples. Incorporating Lactococcus lactis at 107 CFU mL-1 into the samples helped expand the linear range between the cycle threshold values and the concentration of target bacteria. By combining preamplification with inert Lc. lactis, probiotic LAB cells could be detected at single-digit levels in breast milk, while also minimizing the requirements for primer quality and DNA extraction kits. This qPCR-based approach is a reliable tool for studying the potential distribution patterns of LAB in low biomass samples, and it enabled approximately a tenfold increase in the number of breast milk samples in which Lactobacillus and Bifidobacterium species were detected, indicating the ubiquitous Lactobacillus and Bifidobacterium in breast milk.
Keywords: Inert bacteria; Lactic acid bacteria; Low-concentration; Preamplification; Quantitative PCR.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
Laboratory-based molecular test alternatives to RT-PCR for the diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Oct 14;10(10):CD015618. doi: 10.1002/14651858.CD015618. Cochrane Database Syst Rev. 2024. PMID: 39400904
-
[Fast determination of per- and polyfluoroalkyl substances in human serum by cold-induced phase separation coupled with liquid chromatography-tandem mass spectrometry].Se Pu. 2025 Jul;43(7):756-766. doi: 10.3724/SP.J.1123.2024.11028. Se Pu. 2025. PMID: 40610770 Free PMC article. Chinese.
-
Eliciting adverse effects data from participants in clinical trials.Cochrane Database Syst Rev. 2018 Jan 16;1(1):MR000039. doi: 10.1002/14651858.MR000039.pub2. Cochrane Database Syst Rev. 2018. PMID: 29372930 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Probiotics for the prevention of pediatric antibiotic-associated diarrhea.Cochrane Database Syst Rev. 2015 Dec 22;(12):CD004827. doi: 10.1002/14651858.CD004827.pub4. Cochrane Database Syst Rev. 2015. Update in: Cochrane Database Syst Rev. 2019 Apr 30;4:CD004827. doi: 10.1002/14651858.CD004827.pub5. PMID: 26695080 Updated.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

