Genome-wide association and functional annotation analyses reveal candidate genes and pathways associated with various ewe longevity indicators in U.S. Katahdin sheep
- PMID: 40678382
- PMCID: PMC12267044
- DOI: 10.3389/fgene.2025.1600587
Genome-wide association and functional annotation analyses reveal candidate genes and pathways associated with various ewe longevity indicators in U.S. Katahdin sheep
Abstract
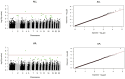
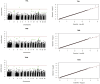
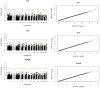
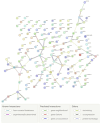
Ewe longevity indicators are complex traits that are lowly heritable, expressed late in life, and sex-limited, making them challenging to include in breeding programs. In this context, genome-wide association studies (GWASs) can provide more information on the complex genetic control of these traits. Therefore, the primary objective of this study was to carry out association analyses for 8 longevity-related traits in 12,734 Katahdin ewes. A total of 126 associations at the chromosome-wide level and 3 at genome-wide level were found. These associations involved 86 single-nucleotide polymorphisms (SNPs) located across 22 chromosomes, with 24 of these SNPs associated with two or more traits. The variants overlapped with genes previously associated with prolificacy (APOH, NLRP9, H3PXD2A, CKB, and HERC4), ovarian follicle pool (GALNT13, TMEM150B, and BRSK1), synthesis and release of reproductive hormones (SULT1B1, LEF1, and EIF5), and early pregnancy events (ITGAV, HADH, ZNFX1, ZSCAN4, EPN1, FBXW8, NOS1, ST3GAL4, and GFRA1). Moreover, genes related to response to stress or pathological conditions (ADCY5, HADH, ATRNL1, LEP, IL11, NLRP9, PRKCG, PRKCA, NEDD4L, FECH, CTNNA3, HECTD1, LRRTM3, and zinc-finger proteins), growth performance (GRID2, MED13L, DCPS, and LEP), and carcass traits (CMYA5 and SETD3) were also implicated. Metabolic pathways such as oxytocin signaling and cardiac-related pathways were enriched. These findings suggest that longevity indicators in Katahdin ewes are highly polygenic traits influenced by a combination of voluntary and involuntary culling reasons. Candidate genes and metabolic pathways influencing reproductive performance and health may play a key role in the functional longevity of Katahdin ewes.
Keywords: genes; genome-wide associate study; lifespan; longevity; ovine; productive life.
Copyright © 2025 Pinto, Lewis, Rocha, Freking, Murphy, Wilson, Nilson, Burke and Brito.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures
Similar articles
-
Heritability estimates and genome-wide association study of methane emission traits in Nellore cattle.J Anim Sci. 2024 Jan 3;102:skae182. doi: 10.1093/jas/skae182. J Anim Sci. 2024. PMID: 38967061 Free PMC article.
-
An Examination of Perceived Stress and Emotion Regulation Challenges as Mediators of Associations Between Camouflaging and Internalizing Symptomatology.Autism Adulthood. 2024 Sep 16;6(3):345-361. doi: 10.1089/aut.2022.0121. eCollection 2024 Sep. Autism Adulthood. 2024. PMID: 39371362
-
Genome-wide association study reveals the QTLs and candidate genes associated with seed longevity in soybean (Glycine max (L.) Merrill).BMC Plant Biol. 2025 Jul 2;25(1):829. doi: 10.1186/s12870-025-06822-1. BMC Plant Biol. 2025. PMID: 40604443 Free PMC article.
-
A meta-analysis of genome-wide association studies to identify candidate genes associated with feed efficiency traits in pigs.J Anim Sci. 2025 Jan 4;103:skaf010. doi: 10.1093/jas/skaf010. J Anim Sci. 2025. PMID: 39847436 Free PMC article.
-
Maternal and neonatal outcomes of elective induction of labor.Evid Rep Technol Assess (Full Rep). 2009 Mar;(176):1-257. Evid Rep Technol Assess (Full Rep). 2009. PMID: 19408970 Free PMC article.
References
-
- Aleri J. W., Lyons A., Laurence M., Coiacetto F., Fisher A. D., Stevenson M. A., et al. (2021). A descriptive retrospective study on mortality and involuntary culling in beef and dairy cattle production systems of Western Australia (1981–2018). Aust. Vet. J. 99, 395–401. 10.1111/avj.13096 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

