Early detection of SARS-CoV-2 variants using genomic surveillance: insights from aircraft wastewater and nasal swabs at Kigali International Airport, Rwanda
- PMID: 40678396
- PMCID: PMC12269423
- DOI: 10.1016/j.ijregi.2025.100678
Early detection of SARS-CoV-2 variants using genomic surveillance: insights from aircraft wastewater and nasal swabs at Kigali International Airport, Rwanda
Abstract
Objectives: The growing threat of emerging infectious diseases necessitates proactive genomic surveillance, particularly, in regions with limited resources and low levels of existing reporting. This study highlights the implementation of a comprehensive genomic surveillance program at the Kigali International Airport and explores the utility of a dual-sample strategy leveraging environmental aircraft wastewater and pooled nasal swab sample types for comprehensive detection and characterization of SARS-CoV-2 lineages being imported into Rwanda.
Methods: Using a combined pooled nasal swab and aircraft wastewater sampling approach resulted in complementary insights in terms of geographic coverage, positivity, and variant characterization.
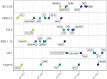
Results: Mutational profiling in source pooled nasal swabs and aircraft wastewater sample data revealed dynamic shifts in mutation prevalence that corresponded with global patterns. Emerging variant JN.1 was detected early in nasal swab data, demonstrating the power of using genomic surveillance as an early warning system.
Conclusions: These results support the feasibility of pathogen surveillance in high-traffic settings and may help drive interest in expanding programs to include pathogens beyond SARS-CoV-2.
Keywords: SARS-Cov2; Surveillance; Traveler genomics.
© 2025 The Author(s).
Conflict of interest statement
CAP, EAL, CWP, and NBC are employed by Ginkgo Bioworks and own Ginkgo Bioworks employee stocks and/or Restricted Stock Units grants.
Figures




References
-
- World Bank open data World Bank Open Data. 2024. https://data.worldbank.org [accessed 01 August 2024]
LinkOut - more resources
Full Text Sources
Miscellaneous

