Novel MRSA-targeting phage MetB16: Genomic features, structural insights, and therapeutic applications
- PMID: 40678418
- PMCID: PMC12266354
- DOI: 10.55730/1300-0152.2746
Novel MRSA-targeting phage MetB16: Genomic features, structural insights, and therapeutic applications
Abstract
Background/aim: Recent reports have indicated that multidrug-resistant strains of S. aureus, including methicillin-resistant strains, may pose a significant threat to public health and global economic stability.
Materials and methods: In this study, we present the isolation and comprehensive characterization of a novel phage, derived from clinically isolated MRSA strains.
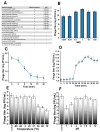
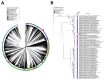
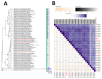
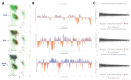
Results: MetB16 exhibited an incubation period of approximately 20 min, a lysis period of around 45 min, and a burst size of 127 Plaque Forming Units (PFU)/cell. The phage demonstrated remarkable biological stability across a pH spectrum of 4.0-9.0 and maintained integrity within a temperature range of 37 and -80 °C. Scanning transmission electron microscopy and phylogenetic analyzes classified MetB16 as belonging to the Triavirus genus, representing a novel species within the Triaviruses. Whole-genome sequencing revealed a 45,295 bp-long genome size with a G + C content of 33.34%. Notably, bioinformatic analyses identified random integration sites within the MRSA genome. Functional annotation of the genome uncovered 72 open reading frames (ORFs), of which 34 encoded hypothetical proteins of unknown function, and these ORFs were associated with phage structure, packaging, host lysis, DNA metabolism, and additional functions. To elucidate the therapeutic potential of temperate phages, detailed structural analyses were conducted on key proteins, including holin, endolysin, and minor tail proteins of MetB16.
Conclusion: This study provides for the first time, the preliminary studies on the biological properties of MetB16 and comprehensive data facilitating an in-depth analysis of the mechanism underlying phage-host interactions, serving as a valuable reference for the evaluation of temperate phages in phage therapy.
Keywords: Methicillin-resistant Staphylococcus aureus (MRSA); modeling phage protein; phage bioinformatics; prophage induction.
© TÜBİTAK.
Conflict of interest statement
Declaration of competing interest: Authors declare that they have no competing interests.
Figures




Similar articles
-
Isolation and characterization of bacteriophages targeting methicillin-resistant Staphylococcus aureus (MRSA) from burn patients and sewage water: a genomic and proteomic study.Int Microbiol. 2025 Aug;28(6):1331-1347. doi: 10.1007/s10123-024-00618-3. Epub 2024 Dec 6. Int Microbiol. 2025. PMID: 39638914
-
Bacteriophage infection drives loss of β-lactam resistance in methicillin-resistant Staphylococcus aureus.Elife. 2025 Jul 10;13:RP102743. doi: 10.7554/eLife.102743. Elife. 2025. PMID: 40637714 Free PMC article.
-
Isolation, characterization and application of noble bacteriophages targeting potato common scab pathogen Streptomyces stelliscabiei.Microbiol Res. 2024 Jun;283:127699. doi: 10.1016/j.micres.2024.127699. Epub 2024 Mar 20. Microbiol Res. 2024. PMID: 38520838
-
Interventions for the eradication of meticillin-resistant Staphylococcus aureus (MRSA) in people with cystic fibrosis.Cochrane Database Syst Rev. 2018 Jul 21;7(7):CD009650. doi: 10.1002/14651858.CD009650.pub4. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2022 Dec 13;12:CD009650. doi: 10.1002/14651858.CD009650.pub5. PMID: 30030966 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Ackermann H, Tremblay D, Moineau S. Long-Term Bacteriophage Preservation. 2004
LinkOut - more resources
Full Text Sources
