Perturbation response scanning of drug-target networks: Drug repurposing for multiple sclerosis
- PMID: 40678478
- PMCID: PMC12268079
- DOI: 10.1016/j.jpha.2025.101295
Perturbation response scanning of drug-target networks: Drug repurposing for multiple sclerosis
Abstract
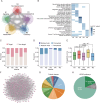
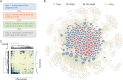
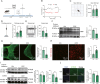
Combined with elastic network model (ENM), the perturbation response scanning (PRS) has emerged as a robust technique for pinpointing allosteric interactions within proteins. Here, we proposed the PRS analysis of drug-target networks (DTNs), which could provide a promising avenue in network medicine. We demonstrated the utility of the method by introducing a deep learning and network perturbation-based framework, for drug repurposing of multiple sclerosis (MS). First, the MS comorbidity network was constructed by performing a random walk with restart algorithm based on shared genes between MS and other diseases as seed nodes. Then, based on topological analysis and functional annotation, the neurotransmission module was identified as the "therapeutic module" of MS. Further, perturbation scores of drugs on the module were calculated by constructing the DTN and introducing the PRS analysis, giving a list of repurposable drugs for MS. Mechanism of action analysis both at pathway and structural levels screened dihydroergocristine as a candidate drug of MS by targeting a serotonin receptor of serotonin 2B receptor (HTR2B). Finally, we established a cuprizone-induced chronic mouse model to evaluate the alteration of HTR2B in mouse brain regions and observed that HTR2B was significantly reduced in the cuprizone-induced mouse cortex. These findings proved that the network perturbation modeling is a promising avenue for drug repurposing of MS. As a useful systematic method, our approach can also be used to discover the new molecular mechanism and provide effective candidate drugs for other complex diseases.
Keywords: HTR2B; Mechanism of action; Multiple sclerosis; Network perturbations.
© 2025 The Author(s).
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures






Similar articles
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Short-Term Memory Impairment.2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Jun 8. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 31424720 Free Books & Documents.
-
Immunomodulators and immunosuppressants for multiple sclerosis: a network meta-analysis.Cochrane Database Syst Rev. 2013 Jun 6;2013(6):CD008933. doi: 10.1002/14651858.CD008933.pub2. Cochrane Database Syst Rev. 2013. PMID: 23744561 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

