NRF2 regulates lipid droplet dynamics to prevent lipotoxicity
- PMID: 40678507
- PMCID: PMC12270665
- DOI: 10.1016/j.isci.2025.112925
NRF2 regulates lipid droplet dynamics to prevent lipotoxicity
Abstract
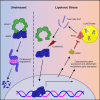
Lipid droplets (LDs) are dynamic organelles comprising a neutral lipid core encapsulated by a phospholipid monolayer. LD structure and function are influenced by a variety of intrinsic and extrinsic signals, and cells alter LD content and distribution to adapt to their environment. Here, we show that LD content increases in response to stabilization of the transcription factor NRF2 under conditions of lipotoxic stress. Notably, NRF2 activity leads to increased expression of the G0S2, a protein that inhibits ATGL, the enzyme responsible for degradation of triacylglycerol and the release of fatty acids from LDs. Importantly, stabilization of NRF2 in the absence of stress is sufficient to increase LD content, and inhibition of ATGL partially rescues the impact of NRF2 deletion on stress-induced ferroptosis. These data support a model in which stress-induced NRF2 stabilization protects cells against lipotoxicity in part through the sequestration of fatty acids in lipid droplets.
Keywords: Biochemistry; Biological sciences; Cell biology.
© 2025 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Mejhert N., Kuruvilla L., Gabriel K.R., Elliott S.D., Guie M.A., Wang H., Lai Z.W., Lane E.A., Christiano R., Danial N.N., et al. Partitioning of MLX-family transcription factors to lipid droplets regulates metabolic gene expression. Mol. Cell. 2020;77:1251–1264.e9. doi: 10.1016/j.molcel.2020.01.014. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

