Deciphering the microbiome-metabolome landscape of an inflammatory bowel disease inception cohort
- PMID: 40679059
- PMCID: PMC12279270
- DOI: 10.1080/19490976.2025.2527863
Deciphering the microbiome-metabolome landscape of an inflammatory bowel disease inception cohort
Abstract
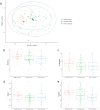
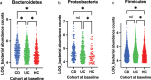
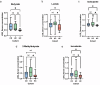
The gut microbiota contribute to the etiopathogenesis of inflammatory bowel disease (IBD), but limitations of prior studies include the use of sequencing alone (restricting exploration of the contribution of microbiota functionality) and the recruitment of patients with well-established disease (introducing potential confounders, such as immunomodulatory medication). Here, we analyze a true IBD inception cohort and healthy controls (HCs) via stool 16S rRNA gene sequencing and multi-system metabolomic phenotyping (using nuclear magnetic spectroscopy and mass spectroscopy), with subsequent integrative network analysis employed to delineate novel microbiota-metabolome interactions in IBD. Marked differences in β diversity and taxonomic profiles were observed both between IBD and HCs, as well as between Crohn's disease (CD) and ulcerative colitis (UC) patients. Multiple between-group metabolomic differences were also observed, particularly with regard to tryptophan-/indole-related metabolites; for example, UC patients had higher levels of serum metabolites including xanthurenic acid (q = 0.0092) and picolinic acid (q = 0.018). Network analysis demonstrated multiple unique interactions in CD compared to HCs with minimal overlap, indicating a loss of 'health-associated' interactions in CD. Compared to HCs, UC patients demonstrated increased pathway activity related to nitrogen and butanoate metabolism, whilst CD patients displayed increased leucine and valine synthesis. Networks from IBD patients overall showed negative correlation with health-specific associations, including an increase in taurine metabolism. Collectively, this work characterizes multiple novel perturbed microbiota-metabolome interactions that are present even at the diagnosis of IBD, which may inform potential future targets to aid diagnosis and direct therapeutic options.
Keywords: Crohn‘s Disease; IBD; Inception cohort; Metabolome; Microbiome; Network analyses; Ulcerative Colitis.
Conflict of interest statement
STR has received conference fees from Pfizer and Vifor with advisory fees from Galapagos. BHM has received consultancy fees from Finch Therapeutics Group, Akebia Therapeutics Inc, and Ferring Pharmaceuticals, and speaker fees from Yakult. SB has received conference fees from Dr Falk and Ferring. RWP declares conference fee support from Ferring. NP declares advisory and/or speaker fees from Abbvie, Allergan, Celgene, Debiopharm, Ferring and Vitor Pharma, and lecture fees from Allergan, Dr Falk Pharma, Janssen, Takeda and Tillotts Pharma. JRM has been paid for consultancy by Cultech Ltd and Enterobiotix Ltd. JLA has received conference fees from Celltrion, Tillots Pharma and Takeda with speaker fees from Abbvie and Johnson & Johnson. HRTW has received financial support to attend conferences from Takeda and has been an advisory board member for Pfizer.
Figures




References
-
- Fyderek K, Strus M, Kowalska-Duplaga K, Gosiewski T, Wedrychowicz A, Jedynak-Wasowicz U, Sładek M, Pieczarkowski S, Adamski P, Kochan P, Heczko, PB. Mucosal bacterial microflora and mucus layer thickness in adolescents with inflammatory bowel disease. World J Gastroenterol. 2009;15(42):5287–5294. doi: 10.3748/wjg.15.5287. - DOI - PMC - PubMed
-
- Imhann F, Vich Vila A, Bonder MJ, Fu J, Gevers D, Visschedijk MC, Spekhorst LM, Alberts R, Franke L, van Dullemen HM, Ter Steege, RWF, Huttenhower, C, Dijkstra, G, Xavier, RJ, Festen, EAM, Wijmenga, C, Zhernakova, A, Weersma, RK. Interplay of host genetics and gut microbiota underlying the onset and clinical presentation of inflammatory bowel disease. Gut. 2018;67(1):108–119. doi: 10.1136/gutjnl-2016-312135. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
